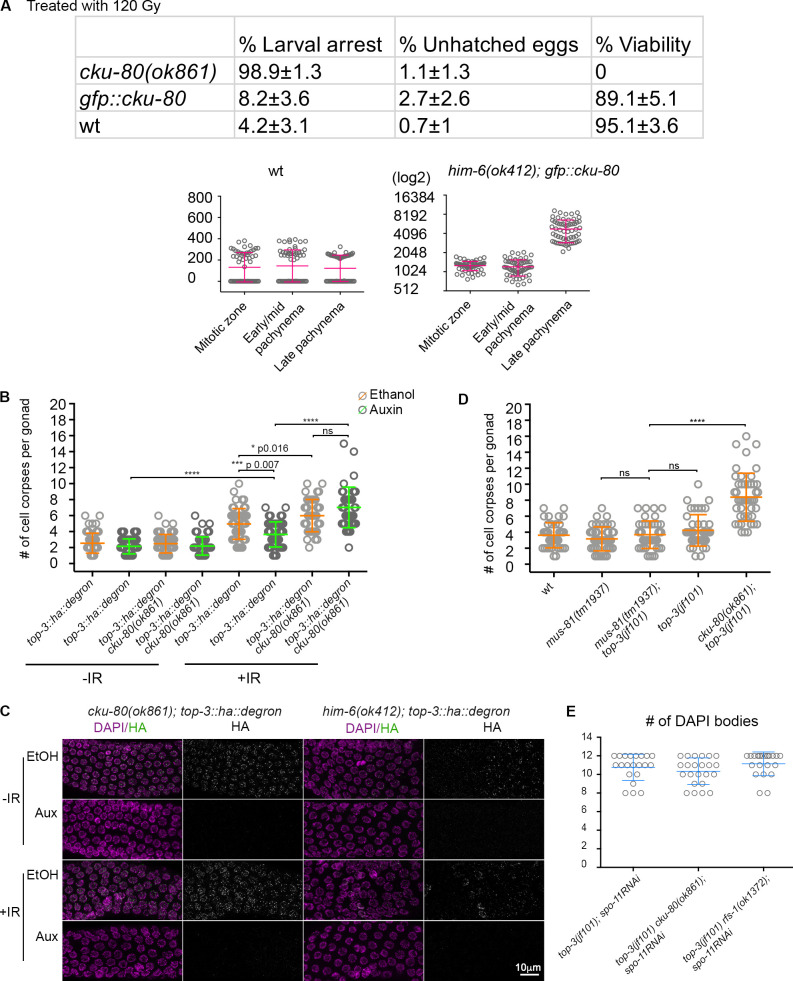
Figure S4.
The GFP::CKU-80 transgene is functional. Related to Fig. 5. (A) Top: Survival after irradiation was assessed by determining larval arrest, the number of unhatched eggs, and viability (i.e., development to adulthood): cku-80, n = 323 embryos; gfp::cku-80, n = 333 embryos; and wt, n = 296 embryos. Note that the tagged version of CKU-80 appears to be functional because the mutant shows little larval arrest, and most progeny are viable. Bottom: Quantification of the fluorescence signal during different prophase I stages in wt (control) and him-6(ok412). The scatter plot shows the mean ± SD. Mean values for the different stages: wt, 132.0–144.7–122.4; him-6(ok412), 1,275–1,224–4,795. (B) Apoptosis quantification in top-3::ha::degron and cku-80(ok861) top-3::ha::degron in the absence (orange) or presence (green) of auxin using SYTO-12 with and without irradiation (120 Gy). Scatter plots indicate the mean ± SD. n = number of gonads scored: top-3::ha::degron (not irradiated) ethanol, 2.5 ± 1.3, n = 67; auxin, 2.2 ± 0.9, n = 58; top-3::ha::degron (after irradiation) ethanol, 5 ± 1.9, n = 51; auxin, 3.6 ± 1.6, n = 49; cku-80(ok861) top-3::ha::degron (not irradiated) ethanol, 2.5 ± 1.2, n = 60; auxin, 2.2 ± 1.1, n = 52; cku-80(ok861) top-3::ha::degron (after irradiation) ethanol, 6 ± 2, n = 44; auxin, 7 ± 2.5, n = 42. (C) Representative cku-80(ok861) top-3::ha::degron and him-6(ok412); top-3::ha::degron nuclei in pachynema, stained with DAPI (magenta) and anti-HA (green) exposed to ethanol (EtOH) or auxin with and without irradiation (120 Gy). (D) Apoptosis quantification using SYTO-12 in the indicated genotypes. Scatter plots indicate the mean ± SD. n = number of gonads scored. wt, 3.6 ± 1.5, n = 39 (quantification as described for Fig. 2 D); mus-81(tm1937), 3.2 ± 1.5, n = 47; top-3(jf101) mus-81(tm1937), 3.7 ± 1.7, n = 42; top-3(jf101), 4.2 ± 1.9, n = 43 (quantification as described for Fig. 2 D); cku-80(ok861) top-3(jf101), 8.4 ± 3, n = 47 (quantification as described for Fig. 6 A). (E) Quantification of DAPI-stained bodies in −1 diakinesis oocytes of different genotypes treated with spo-11RNAi. Error bars indicate the mean ± SD. n = number of diakinesis nuclei scored: top-3(jf101); spo-11RNAi, 10.8 ± 1.4, n = 21; top-3(jf101) cku-80 (ok861); spo-11RNAi, 10.35 ± 1.4, n = 23; and top-3(jf101) rfs-1(ok1373); spo-11RNAi, 11.1 ± 1.3, n = 21. Aux, auxin; wt, wild-type.