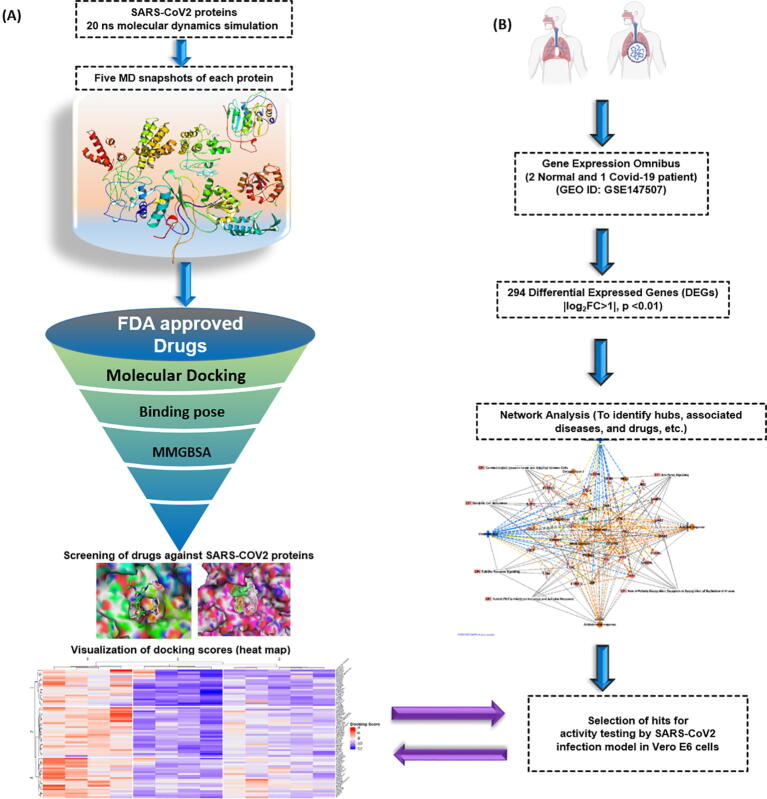
Fig. 1.
Outline of the repurposing work. (A) Nine SARS-CoV2 proteins structures were obtained from either PDB or modeled. The protein structures were subjected to 20 ns MD run (total 180 ns) to explore the flexibility of the binding site. Snapshots were generated at equal time points during MD simulation for each protein. The FDA approved drug library was docked in the generated snapshots using Glide. The MMGBSA score was also calculated for each of the ligand in individual snapshots. (B) Transcriptomics data from SARS-CoV2 infected and normal human samples identified significantly differentially expressed genes (|log2FC > 1|, p-value < 0.01) as a result of infection. A protein–protein-interaction network was created using these genes. They were also analyzed for their involvement in biological pathways using Ingenuity Pathways Analysis (IPA). The drug gene interactions were also analysed and molecules were tested using SARS-CoV2 infection model in Vero E6 cells for antiviral activity.