Figure 1: Orthogonal analyses identify the 2OG-dependent dioxygenase JMJD6 as a potential regulator of AR-V7.
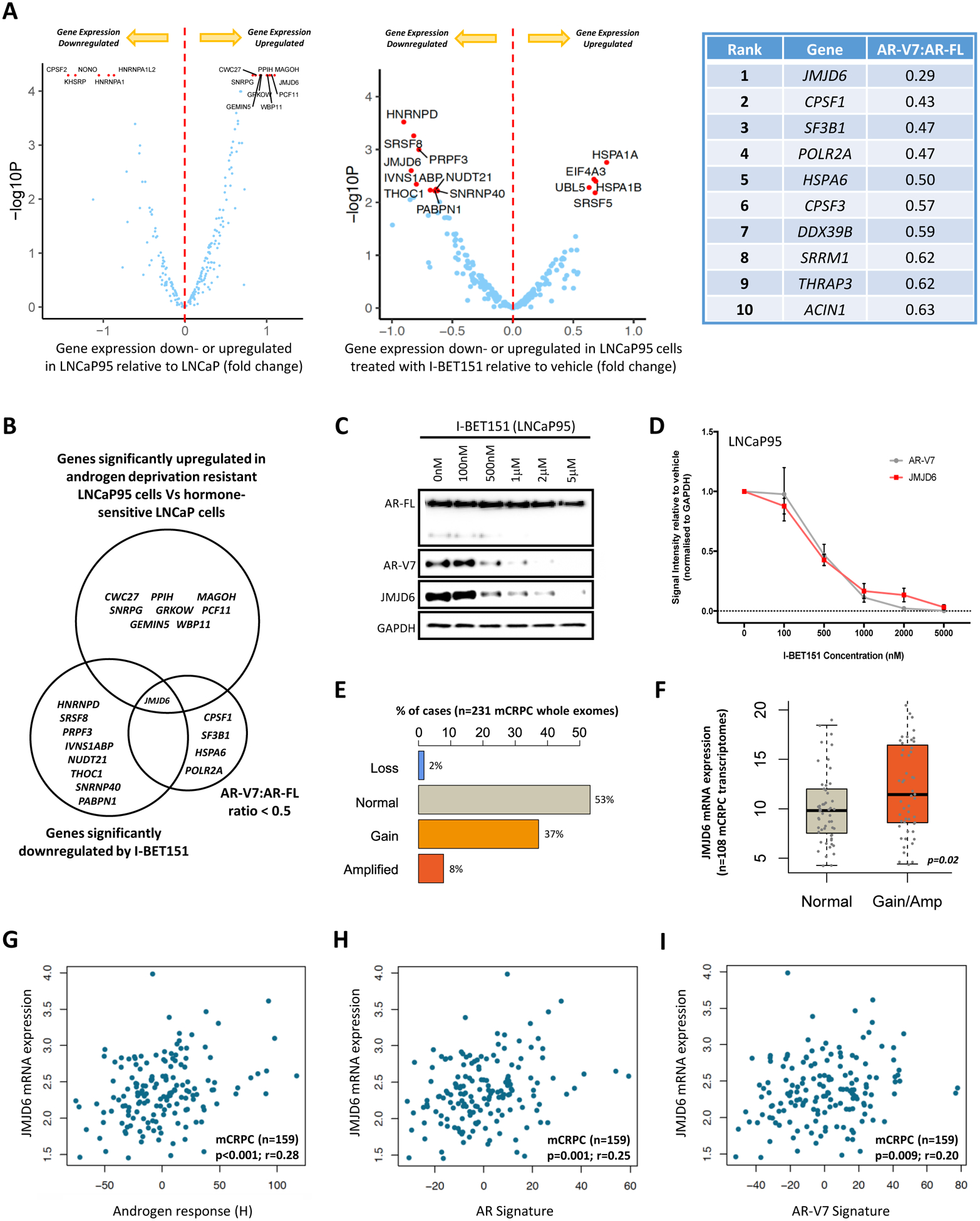
(A) Volcano plots illustrating differential mRNA expression of 315 genes relating to the spliceosome (spliceosome related gene set; supplementary table 6), as determined by RNA-seq, between hormone-sensitive LNCaP (no AR-V7 protein) and androgen deprivation resistant LNCaP95 (detectable AR-V7 protein) prostate cancer (PC) cell lines, and LNCaP95 PC cells treated with either a BET inhibitor (I-BET151) or vehicle (DMSO 0.1%). Blue dots represent genes with baseline expression (FPKM) greater than the median expression level of all 315 genes at baseline across both experiments. Top 15 genes most differentially expressed (FPKM) in each experiment (up- or down-regulated) indicated by red dots. Top 10 hits identified in targeted siRNA screen shown in accompanying table; all 315 genes in the spliceosome related gene set were individually inhibited by siRNA in 22Rv1 and LNCaP95 PC cell lines. Changes in AR-V7 protein levels relative to AR-FL were quantified by western blot (WB) densitometry. AR-V7 downregulation averaged across both cell lines with genes ranked in order of the degree of AR-V7 downregulation relative to AR-FL. (B) Venn diagram amalgamating RNA-seq analyses with siRNA screen results. Genes of interest pre-defined as being upregulated in LNCaP95 cells relative to LNCaP cells, downregulated following BET inhibition, and associated with a > 50% reduction in AR-V7 protein expression (WB) relative to AR-FL following siRNA knockdown. JMJD6 was the only gene to meet all three criteria. (C) WB demonstrating that I-BET151 treatment (48 hours) in LNCaP95 PC cells downregulates both AR-V7 and JMJD6 protein expression in a dose-dependent manner. Single representative WB shown from four separate experiments. (D) Densiometric quantification of JMJD6 (red line) and AR-V7 (grey line) protein levels (n=4; densitometry for each biological replicate normalized to GAPDH and vehicle). Demonstrates that protein levels of both JMJD6 and AR-V7 decrease in a dose-dependent manner following BET inhibition with I-BET151. (E) Whole exome analysis (n=231) shows alterations of the JMJD6 gene locus were detected in 47% of mCRPC biopsies (SU2C/PCF cohort), with these being predominately gains (37%; n=86) or amplifications (8%; n=18). (F) Whole exome analysis of mCRPC patients with matched transcriptome data from SU2C/PCF cohort (n=108) shows that JMJD6 copy number gain and amplification (Amp) associated with an increase in JMJD6 mRNA expression in mCRPC biopsies compared to samples without JMJD6 copy number gain/amplification (p=0.02; Wilcoxon test). (G-I) Scatter plots of transcriptome analysis in 159 mCRPC biopsies (SU2C/PCF cohort) showing correlations between JMJD6 mRNA expression and (G) androgen response (Hallmark; H), (H) AR signature (derived from 43 AR regulated transcripts) and (I) AR-V7 signature (derived from 59 genes associated with AR-V7 expression in mCRPC). JMJD6 mRNA expression shown as log FPKM. r-values and p-values are shown and were calculated using Spearman’s correlation.
