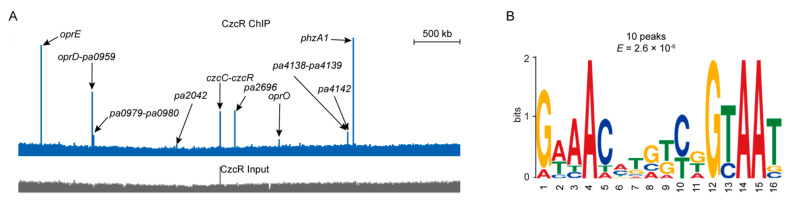
Figure 2.
Identification of CzcR targets. (A) A representative image of CzcR ChIP-seq result using Integrated Genome Viewer. ChIP sample (blue) and the input control sample (grey) were shown. Arrows showing the ChIP-seq peaks located in the promoters of the indicated genes. (B) The most significant motif was generated by the MEME tool using 101 bp centered on the peak summits of the top 10 peak sequences (Data set 1, sheet 8), with minimum width as 3 and other parameters as default. The height of each letter represents the frequency of each base in different positions in the consensus sequence. The 16 nt motif was present in all the 10 tested CzcR binding sites with an E-value of 2.6 × 10-6.