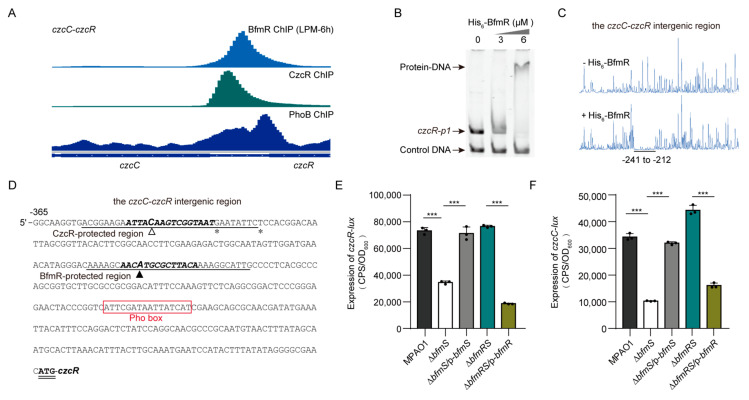
Figure 3.
BfmR binds to and inhibits the promoter activity of czcR and czcC. (A) Pattern of BfmR, CzcR, and PhoB ChIP-seq peaks for the czcC-czcR intergenic region. The PhoB ChIP data were obtained from NCBI Gene Expression Omnibus (GEO) database under accession number GSE128430 [58]. (B) EMSA showing that the N-terminal His6-tagged BfmR (His6-BfmR) binds to the promoter of czcR (i.e., czcR-p1). DNA fragments C-bfmR serves as a negative control. (C) Electropherograms show the protection pattern of the czcC-czcR intergenic region after digestion with DNase I following incubation in the absence (-) and presence (+) of His6-BfmR (6 μM). The protected regions (relative to the start codon of czcR) are underlined. (D) czcR promoter sequence with a summary of the results of DNase I footprint assays and ChIP-seq experiments. The CzcR- and BfmR-protected regions are underlined as indicated and the asterisk shows the DNase I hypersensitivity site described in Figure S2E. Hollow and solid triangle indicates the location of the summits of CzcR- and BfmR ChIP-seq peaks, respectively. The potential Pho box [59] is highlighted by square frame, and the starting codon (ATG) of czcR is in bold and double underlined. Sequences that match the MEME motif of CzcR (see in Figure 2B) and BfmR (see in Figure 1C) are in bold and italic. (E,F) The promoter activity of czcR (in E) and czcC (in F) in wild type (WT) MPAO1 and its derivatives grown in MM supplemented with 50 μM ZnCl2 at 37 °C for 6 h. Data points are shown in black dots, and results represent means ± SD (n = 3 biological replicates; *** p < 0.001, Student’s two-tailed t-test). MPAO1, ΔbfmS, and ΔbfmRS harboring an empty pAK1900 vector as a control; p-bfmS and p-bfmR respectively denote pAK1900-bfmS and pAK1900-bfmR (Table S1).