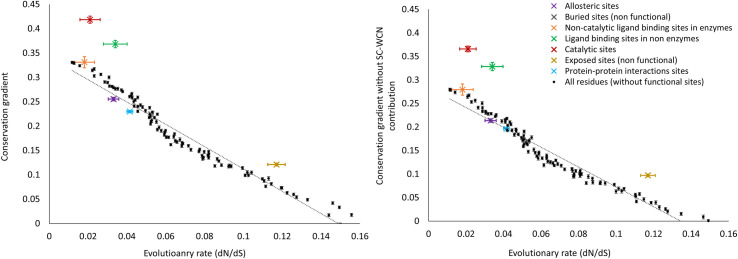
Fig 3. Conservation gradient induced from residues is linearly (negatively) correlated with their evolutionary rate (dN/dS), with catalytic site residues inducing stronger conservation gradient than expected by the linear trend.
(A) Average conservation gradient (calculated as the average Pearson correlation between conservation of residues and their distance from a site) as a function of average evolutionary rate (dN/dS) for all yeast protein residues binned according to their annotated conservation rank into 100 equally spaced bins as well as the average conservation gradients of different types of functional sites. (B) Average conservation gradient without the relative contribution of SC-WCN as a function of average evolutionary rate (dN/dS) for all yeast protein residues binned according to their annotated conservation rank into 100 equally spaced bins as well as the average conservation gradients without the relative contribution of SC-WCN of different types of functional sites.