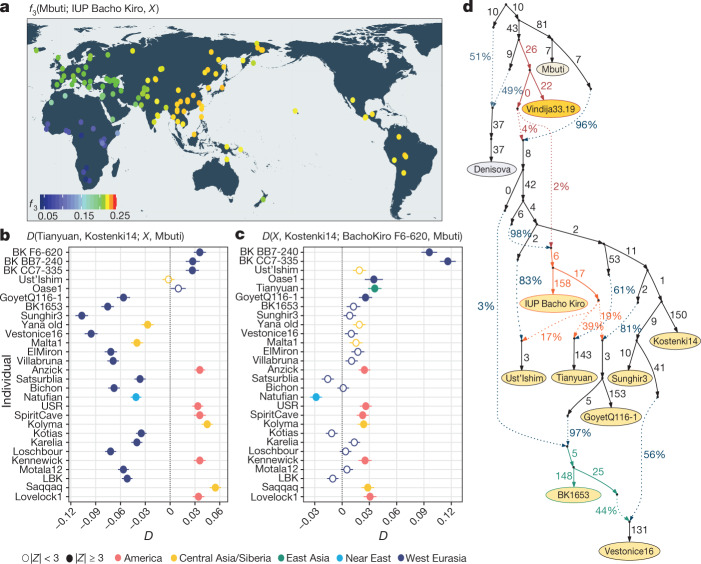
Fig. 2. Population affinities of the IUP Bacho Kiro Cave individuals.
a, Allele sharing (f3) between the IUP Bacho Kiro Cave individuals and present-day populations (X) from the Simons Genome Diversity Project (SGDP)31 after their separation from an outgroup (Mbuti) (calculated as f3(Mbuti; IUP Bacho Kiro, X). Warmer colours on the map48 correspond to higher f3 values (higher shared genetic drift). b, IUP Bacho Kiro Cave individuals share significantly more alleles (proportions of allele sharing or D values plotted on x axis) with the roughly 40,000-year-old Tianyuan individual13 than with the approximately 38,000-year-old Kostenki14 individual29,30. Calculated as D(Tianyuan, Kostenki14; X, Mbuti). c, F6-620 shares significantly more alleles with the Oase17 and GoyetQ116-129 individuals, ancient Siberians and Native American individuals than with the Kostenki14 individual. Calculated as D(X, Kostenki14; F6-620, Mbuti). b, c, Filled circles indicate a significant value (|Z| ≥ 3); open circles, |Z| < 3. Whiskers correspond to 1 s.e. calculated across all autosomes (1,813,821 SNPs) using a weighted block jackknife28 and a block size of 5 Mb. BK, Bacho Kiro. d, Admixture graph relating Bacho Kiro Cave individuals and ancient humans older than 30 kyr bp. This model uses 281,732 overlapping SNPs in all individuals and fits the data with a single outlier (Z = 3.22). Ancient non-Africans (yellow circles), Vindija 33.19 Neanderthal (orange), Denisovan (grey) and present-day African individuals (light yellow circle) are shown. Admixture edges (dotted lines) show the genetic component related to Neanderthals (red), to the IUP Bacho Kiro Cave individuals (orange) and to BK1653 (green). Numbers on solid branches correspond to the estimated drift in f2 units of squared frequency difference; labels on dotted edges give admixture proportions.