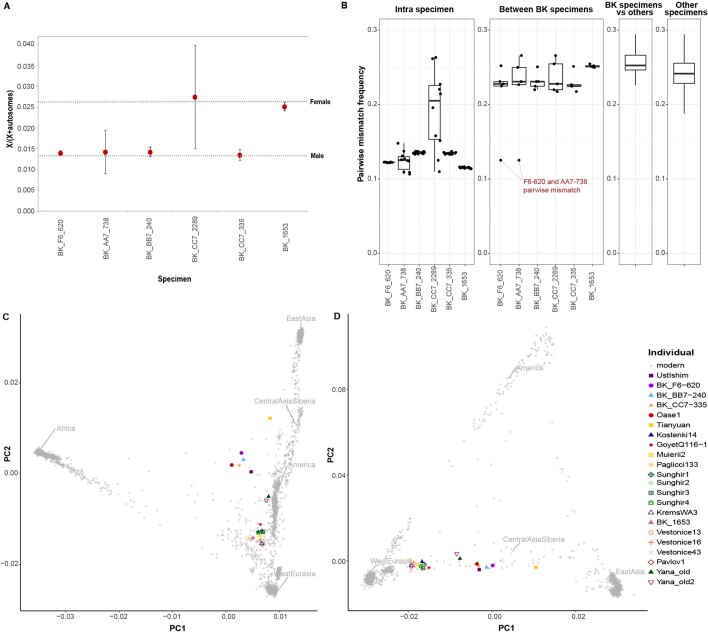
Extended Data Fig. 2. Sex determination, pairwise mismatch rate between specimens and principal component analyses (PCAs).
a, Sex determination for Bacho Kiro Cave specimens. Only fragments that showed C-to-T substitutions in the first three and/or last three positions and overlapping 2200k Panel SNPs were used for this analysis (for the number of deaminated fragments per specimen, see Supplementary Table 2.8). The expected ratios of X to (X + autosomal) fragments for a female and a male individual are depicted as dashed lines, and circles correspond to the calculated values for each of the Bacho Kiro Cave specimens. Whiskers correspond to 95% binomial confidence intervals. b, Pairwise mismatch rate between different libraries from the same specimen (intra-specimen), between different Bacho Kiro Cave specimens (inter-specimen) and between other ancient modern humans older than 30,000 cal. bp. The boxplots were drawn using the summary statistics geom_stat from the R-package ggplot; lower and upper hinges, first and third quartiles; whiskers, maximum value of 1.5× the interquartile range; centre line, median. SNPs across all autosomes of the 2200k Panel were used for the calculations (number of SNPs (nsnps) = 2,056,573). c, A PCA of 2,970 present-day humans genotyped on 597,573 SNPs with 22 ancient individuals older than 30,000 cal. bp projected onto the plane. d, A PCA of 1,444 present-day Eurasian and Native American individuals genotyped on 597,573 SNPs with 22 ancient individuals older than 30,000 cal. bp projected onto the plane. c, d, Grey dots denote present-day human genomes.