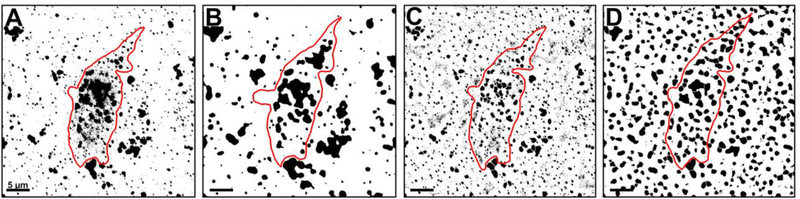
Figure 4.
ImageJ was unable to replicate the image analysis performed in Imaris, despite application of similar methodology. A. Representative image of a 3D reconstructed BFCN (outlined in red) after thresholding in ImageJ. Early endosomes (black) have merged to form large masses making analysis equivocal. B. Application of a Gaussian filter and background subtraction akin to the Imaris “Surface” creation step reduced the number of staining artifacts and salt-and-pepper noise classified as foreground pixels during thresholding. Clusters of endosomes were still difficult to resolve. C. Application of Otsu’s local thresholding, similar to the Imaris “Spots” creation step, reduced excessive noise around the periphery of endosomal clusters and enabled more precise application of watershed. Early endosomes were more reminiscent of their original shape prior to the global thresholding originally performed in (A), however image noise was still high. D. Application of all 4 techniques produced an over processed image that was still difficult to analyze. Scale bar A-D: 5 μm.