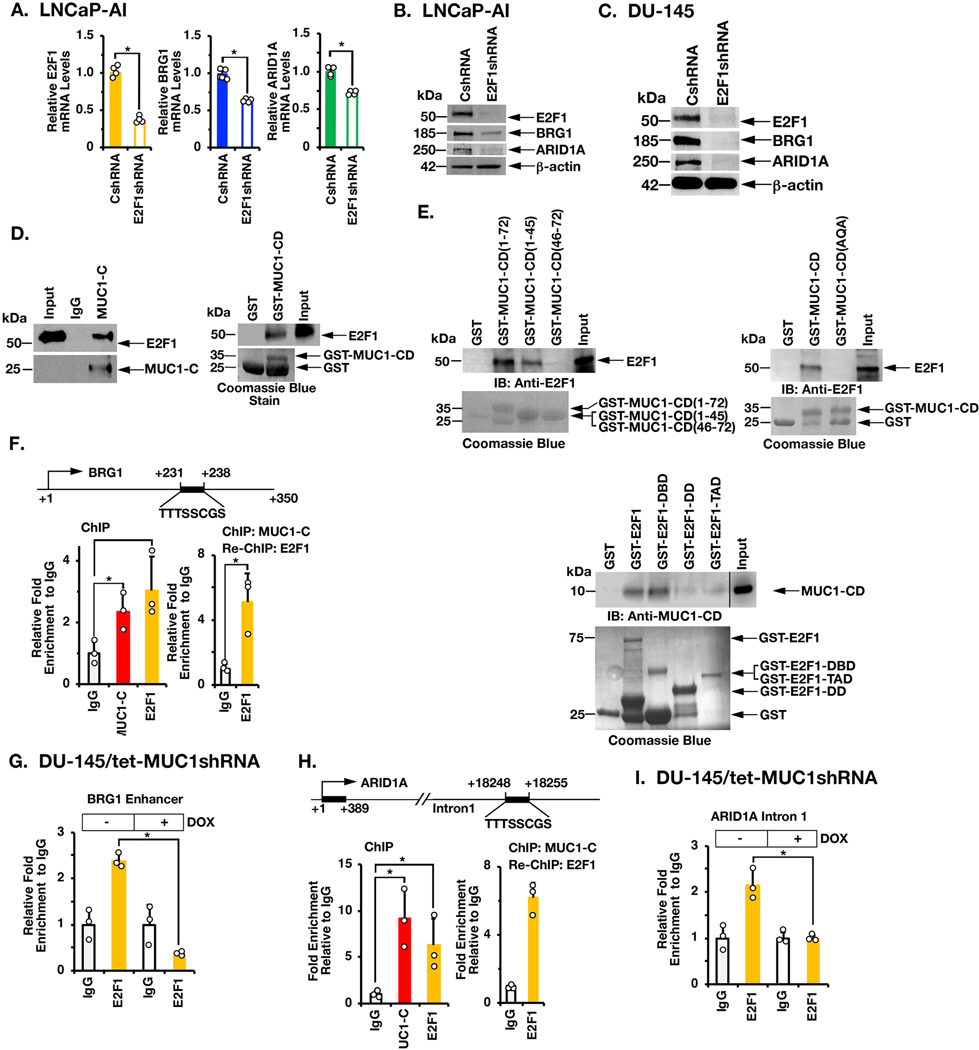
Figure 2. MUC1-C interacts with E2F1 in driving BRG1 and ARID1A expression.
A. LNCaP-AI/CshRNA and LNCaP-AI/E2F1shRNA cells were analyzed for the indicated mRNA levels by qRT-PCR. The results (mean±SD of 4 determinations) are expressed as relative mRNA levels compared to that in control cells (assigned a value of 1). B. Lysates from LNCaP-AI/CshRNA and LNCaP-AI/E2F1shRNA cells were immunoblotted with antibodies against the indicated proteins. C. Lysates from DU-145/CshRNA and DU-145/E2F1shRNA cells were immunoblotted with antibodies against the indicated proteins. D. Nuclear lysates from LNCaP-AI cells were incubated with anti-MUC1-C or a control IgG. The input and precipitates were analyzed by immunoblotting with anti-E2F1 and anti-MUC1-C (left). Purified E2F1 was incubated with GST or GST-MUC1-CD(FL; 1–72) bound to glutathione beads. Adsorbates were immunoblotted with anti-E2F1 (right). Input of the GST proteins was assessed by Coomassie blue staining. E. Purified E2F1 was incubated with GST, GST-MUC1-CD(FL; 1–72) or the indicated GST-MUC1-CD fragments bound to glutathione beads (left). Purified E2F1 was incubated with GST, GST-MUC1-CD(FL; 1–72) or the GST-MUC1-CD(CQC→AQA) mutant bound to glutathione beads (right). Adsorbates were immunoblotted with anti-E2F1. Purified MUC1-CD was incubated with GST, GST-E2F1(FL, 1–437), GST-E2F1(DBD; aa 1–191), GST-E2F1(DD; aa 192–284) and or GST-E2F1(TAD; aa 285–437) bound to glutathione beads (lower). Adsorbates were immunoblotted with anti-MUC1-CD. Input of the GST proteins was assessed by Coomassie blue staining. F. Schema of the BRG1 enhancer region with positioning of the putative E2F1 binding motif. Soluble chromatin from DU-145 cells was precipitated with anti-MUC1-C, anti-E2F1 or a control IgG (left). Soluble chromatin was precipitated with anti-MUC1-C (ChIP) and then reprecipitated with anti-E2F1 or a control IgG (re-ChIP) (right). G. DU-145/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days. Soluble chromatin was precipitated with anti-E2F1 or a control IgG. The DNA samples were amplified by qPCR with primers for the BRG1 enhancer region. The results (mean±SD of 3 determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1). H. Schema of the ARID1A intron 1 region with positioning of the putative E2F1 binding motif. Soluble chromatin from DU-145 cells was precipitated with anti-MUC1-C, anti-E2F1 or a control IgG (left). Soluble chromatin was precipitated with anti-MUC1-C (ChIP) and then reprecipitated with anti-E2F1 or a control IgG (re-ChIP) (right). I. DU-145/tet-MUC1shRNA cells were treated with vehicle or DOX for 7 days. Soluble chromatin was precipitated with anti-E2F1 or a control IgG. The DNA samples were amplified by qPCR with primers for the ARID1A intron 1 region. The results (mean±SD of three determinations) are expressed as the relative fold enrichment compared to that obtained with the IgG control (assigned a value of 1).