FIGURE 2.
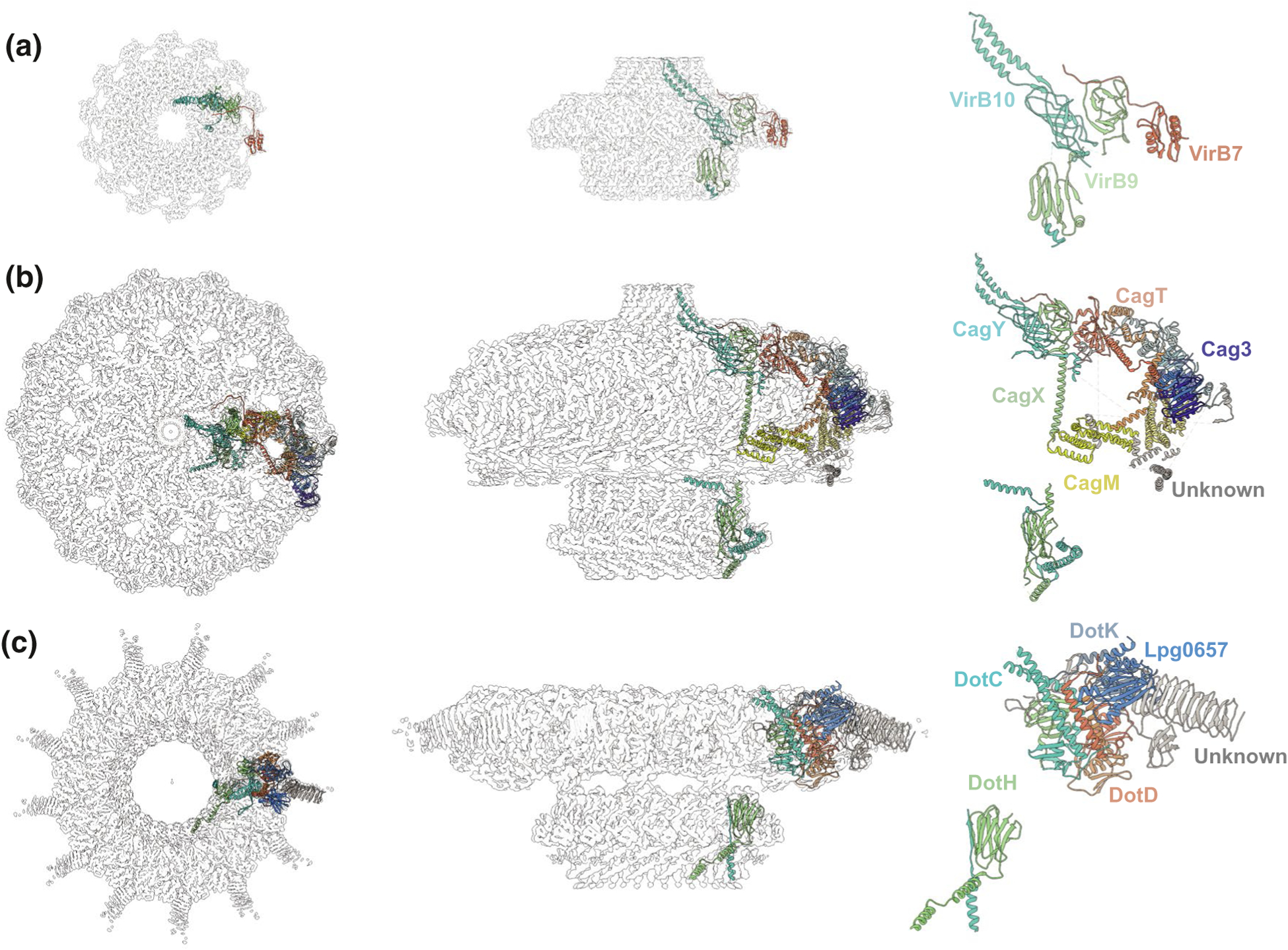
High-resolution CryoEM structures from different T4SS outer membrane complexes. (a) The X. citri CryoEM density map (EMDB-0089) resolved at 3.28 Å with 14-fold symmetry; the map is fitted with an asymmetric unit from PDB model 6gyb. (b) The H. pylori CryoEM density maps of the OMC and PR subdomains (OMC:EMD-22081 and PR:20021) resolved at 3.4 Å with 14-fold symmetry and 17-fold symmetry, respectively; the maps are fitted with asymmetric units from PDB models 6×6j and 6×6s. C) The L. pneumophila OMC disk (EMD-22068) resolved at 3.5 Å with 13-fold symmetry and the PR (EMD-22069) resolved at 3.7 Å with 18-fold symmetry, the maps are fitted with asymmetric units from PDB models 6×62 and 6×64. Locations of confirmed subunits are shown, with similar color-coding for the VirB7, VirB9, and VirB10 homologs or orthologs in each system
