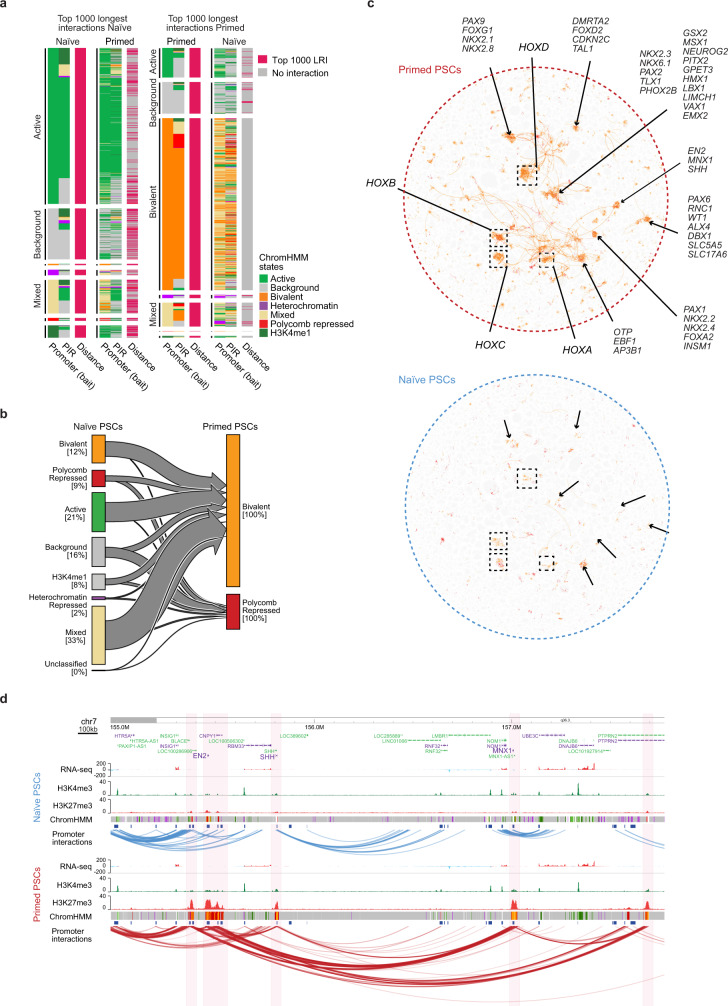
Fig. 3. Long-range promoter interactions are associated with bivalent chromatin in primed human PSCs.
a Heatmaps show the chromatin states of the promoters and promoter-interacting regions (PIRs) that are connected by the longest interactions in naive (left) and primed (right) PSCs. Each heatmap is ordered and divided by the chromatin state of the promoter HindIII fragment. b Plot shows the chromatin states of HindIII fragments in naive PSCs that transition into the bivalent or Polycomb-associated states in primed PSCs. The percentages shown are within each state. c Visualising all bivalent and Polycomb-associated interactions on the network graph highlights several interaction clusters particularly in primed PSCs (upper) that contain many developmental genes. The clusters, together with the interactions, are largely absent in naive PSCs (lower). d Genome browser view in naive (upper) and primed (lower) PSCs of a ~2.5 Mb region that contains several developmental genes including EN2, SHH and MNX1. Significant promoter interactions are shown as blue and red arcs. ChIP-seq (H3K4me3 and H3K27me3) and strand-specific RNA-seq tracks are shown. Chromatin states for each genomic region were defined by ChromHMM46 using ChIP-seq data (active chromatin, light green; H3K4me1-only chromatin, dark green; bivalent chromatin, orange; Polycomb-associated, red; heterochromatin, purple; background, grey).