Fig. 1. Trio-free phased diploid genome assembly using Strand-seq (PGAS).
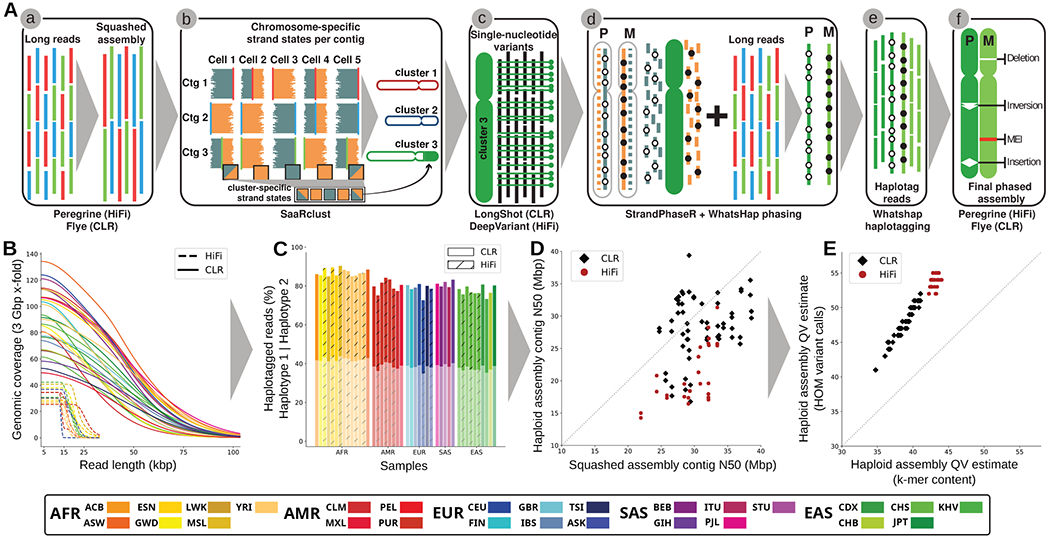
(A) A schematic of the PGAS pipeline (3): (a) generation of a non-haplotype-resolved (“squashed”) long-read assembly; (b) clustering of assembled contigs into “chromosome” clusters based on Strand-seq Watson/Crick signal; (c) calling of single-nucleotide variants (SNVs) relative to the clustered squashed assembly; (d) integrative phasing combines local (SNV) and global (Strand-seq) haplotype information for chromosome-wide phasing; (e) tagging of input long reads by haplotype; (f) phased genome assembly based on haplotagged long reads and subsequent variant calling (18). (B) Genomic coverage (y-axis) as a function of the long-read length (x-axis). (C) Fraction of reads that can be assigned (“haplotagged”) to either haplotype 1 (semitransparent) or haplotype 2 for HiFi (hatched) and CLR (solid) datasets. (D) Contig-level N50 values for squashed (x-axis) and haploid assemblies (y-axis) for CLR (black diamonds) and HiFi (red circles) samples. (E) Haploid assembly QV estimates computed from unique and shared k-mers (x-axis) based on homozygous Illumina variant calls (y-axis). Samples colored according to the 1000GP population color scheme (15) with exception of the added Ashkenazim individual NA24385/HG002 (Coriell family ID 3140) (ASK/dark blue).
