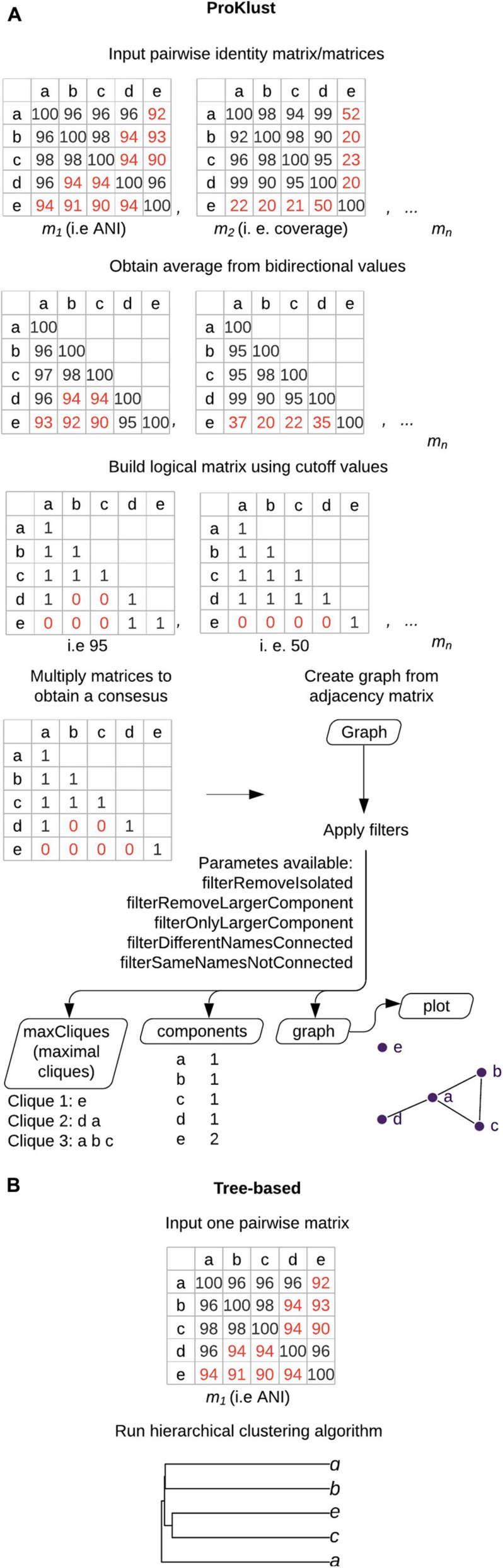
FIGURE 2.
Graph-based clustering using ProKlust compared to hierarchical clustering. (A) The average of each pair from the pairwise input. matrix/matrices is/are obtained. A Boolean matrix/matrices is/are obtained according to the cut-off values chosen by the user. If more than one matrix is used as input, the final generated matrix is obtained by multiplying the elements of the matrices. A graph is formed by connecting the nodes which present the positive values. In this example, nodes correspond to genomes and edges correspond to ANI ≥95% with coverage alignment ≥50%. The data could be filtered to retain components containing more than one species name or unconnected nodes containing the same species names. In addition, filters to remove isolated nodes (“filterRemoveIsolated”) or the largest component (“filterOnlyLargerComponent”) are also available. The tool generates four types of outputs: (i) the maximal cliques on “maxCliques,” which is the largest subset of nodes in which each node is directly connected to every other node in the subset i.e., all the possible species groups that could be delimited in the graph, which could result in groups having genomes in common; (ii) “components” that contains the isolated nodes or groups formed of complete graphs; (iii) “graph,” an igraph object graph, that can be further handled by the user; and (iv) the “plot,” where the final graph could be visualized. (B) Overview of the hierarchical-based clustering approach. These approaches return tree-shaped diagrams with non-overlapping clusters.