TABLE 1.
Tengu homologs used in this study.
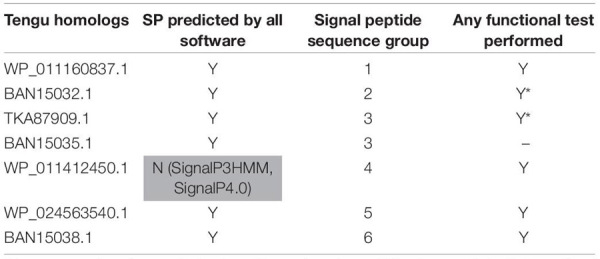 |
The second column indicates if a signal peptide is predicted by all of the SignalP3HMM, SignalP4, SignalP4.1sensitive, Phobius_SP, Phobius_SP_TM software (mention “Y” = yes). If not, cells were highlighted in gray (for better readability) with a “N” (no), and predictors that did not produce a positive prediction are mentioned. The third column shows the signal peptide group based on predictions of SignalP4.1sensitive (identical numbers are attributed to identical sequences). Finally, the last column mentions the presence of available evidence of effector activity (any functional test) from literature with the mention “Y”; * cases where the sequence of the mature protein is identical to another accession for which there is available data (see Supplementary Tables 1–3 for references).
