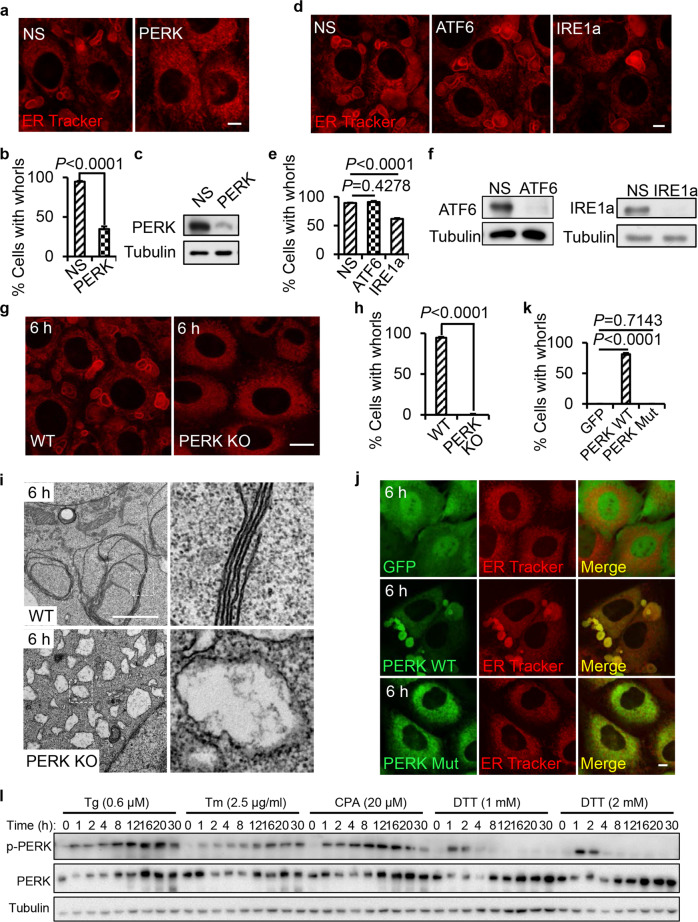
Fig. 2. ER whorl formation is dependent on PERK activation.
a NRK cells were transfected with nonspecific (NS) or PERK siRNA. Cells were treated with Tg for 6 h, and then stained with ER-Tracker Red and visualized by confocal microscopy. Scale bar, 5 μm. b Cells from a were quantified for ER whorls (n = 3 independent experiments; >100 cells were assessed per independent experiment). Data represent means ± SE. c PERK knockdown efficiency in cells from a was determined by western blot. d NRK cells were transfected with NS, ATF6, or IRE1a siRNA. Cells were treated with Tg for 6 h, and then stained with ER-Tracker Red and visualized by confocal microscopy. Scale bar, 5 μm. e Cells from d were quantified for ER whorls (n = 3 independent experiments; >100 cells were assessed per independent experiment). Data represent means ± SE. f Knockdown efficiency in cells from d was determined by western blot. g A PERK KO cell line was generated by CRISPR-Cas9. WT and PERK KO cells were treated with Tg for 6 h, and then stained with ER-Tracker Red and visualized by confocal microscopy. Scale bar, 10 μm. h Cells from g were quantified for ER whorls (n = 3 independent experiments; >100 cells were assessed per independent experiment). Data represent means ± SE. i Representative TEM micrographs of cells from g. Scale bar, 2 μm. Regions outlined with white dashed lines are magnified on the right. j PERK KO cells transfected with GFP, GFP-PERK, or GFP-PERK-Mut (K622A) were treated with Tg for 6 h and then observed by Opera Phenix microscopy with 60× confocal mode. Scale bar, 5 μm. k Cells from j were quantified for ER whorls (n = 3 independent experiments; >100 cells were assessed per independent experiment). Data represent means ± SE. l NRK cells were treated with Tg (0.6 μM), Tm (2.5 μg/mL), CPA (20 μM), DTT (1 mM) or DTT (2 mM) for the indicated times and analyzed by western blot. P values were calculated using a two-tailed, unpaired t-test (b, e, h, k).