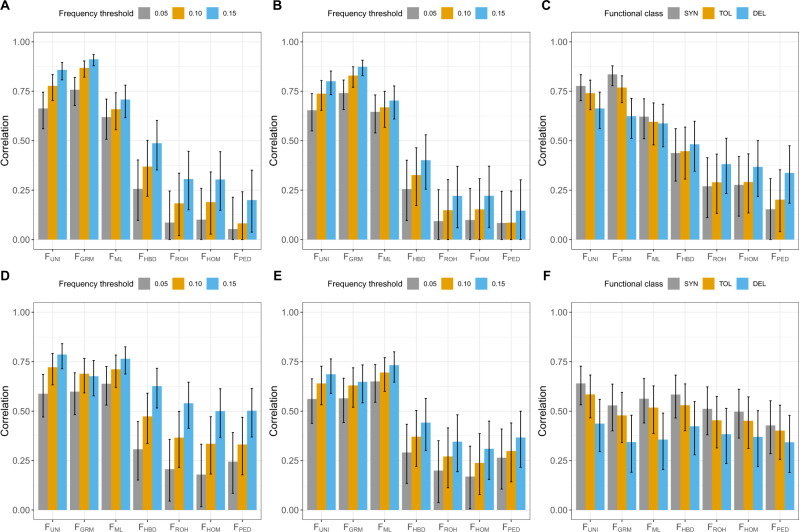
Fig. 2. Correlation coefficients between individual inbreeding measures estimated with 37,675 SNPs and homozygous mutation load (HML) obtained from the whole-genome sequence data in 145 individuals.
HML was computed using alternate (a, b, c) and private alternate (d, e, f) alleles. a and d Correlation with HML estimated with allele frequency thresholds of 0.05, 0.10 and 0.15. b and e Correlation with weighted HML estimated with allele frequency thresholds of 0.05, 0.10 and 0.15. c and f Correlation with HML estimated with synonymous (SYN), tolerated (TOL) and deleterious (DEL) missense variants and using an allele frequency threshold of 0.15. The error bars represent the 95% confidence intervals and are truncated at 0.