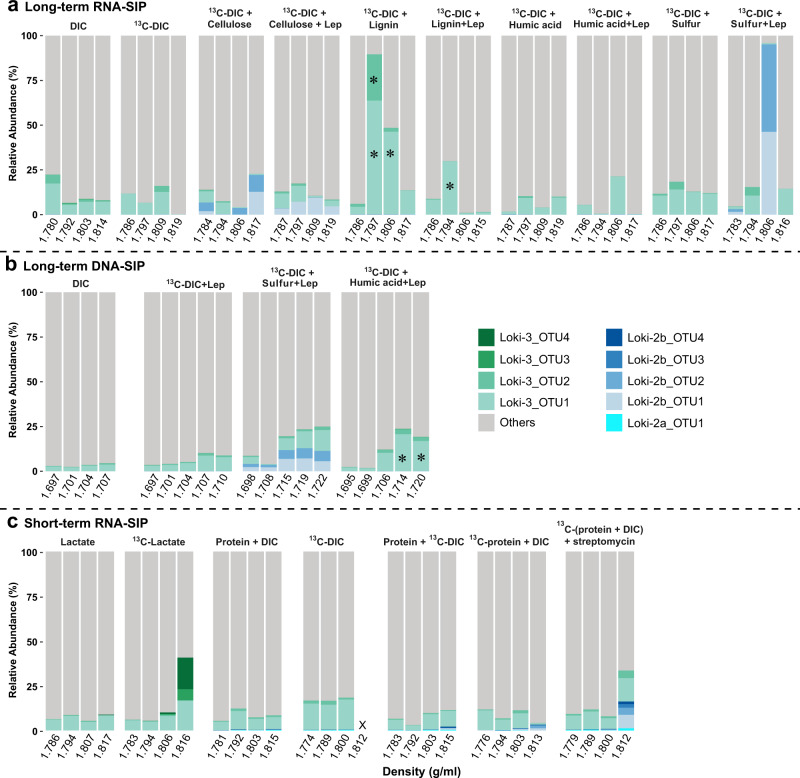
Fig. 2. Total sum scaling charts of Lokiarchaeota abundances of archaeal 16S rRNA gene sequences from selected “light” and “heavy” gradient fractions.
a Long-term RNA-SIP samples amended with 13C-DIC, b Long-term DNA-SIP incubations amended with 13C-DIC, c Short-term RNA-SIP samples from lactate and protein incubations. Differences in x-axis scales between RNA and DNA-SIP are due to the different gradient media used (CsTFA vs CsCl, respectively). For RNA-SIP, pairs of fractions (fractions 4 and 5, 6 and 7, 8 and 9, 10 and 11) were combined for Illumina sequencing, whereas individual fractions were used for DNA-SIP. Density was indicated as the average density of combined fractions for RNA-SIP samples. Due to density differences between RNA and DNA, the threshold density fractions to delineate 13C-labelled nucleic acids differ between RNA (>1.797 g/ml) and DNA (>1.702 g/ml). “X” indicates that cDNA synthesis failed because of insufficient amount of RNA in these fractions. For the Loki-2 OTUs which were not detectable in controls, label incorporation activity was detected by their presence in heavy fractions. An asterisk indicates inter-gradient increase of Loki-3 OTUs (see Fig. S3 for intra-gradient assessment; both approaches were in agreement). DNA with densities >1.71 g/ml was not obtained from DIC incubations. Lep lepidocrocite. DIC dissolved inorganic carbon, i.e. bicarbonate.