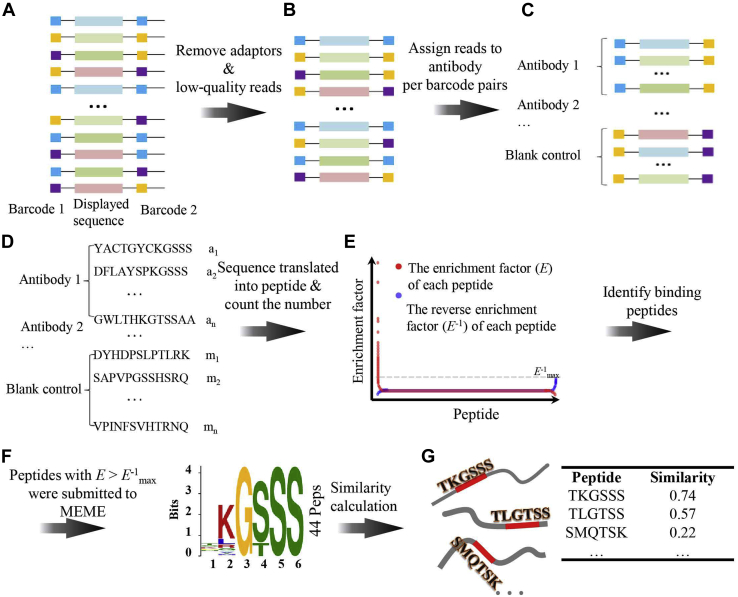
Fig. 2.
Workflow of data processing.A, the structure of the construct for NGS. The solid black lines represent fixed sequences and colored boxes represent varied sequences. B, the filtered NGS data. C, illustration of the reads assigned to antibodies. D, illustration of the peptides assigned to antibodies. E, the algorithm used to obtain binding peptides. For each peptide, both E (red dots) and E−1 (blue dots) are plotted. Only peptides with red dots above the dashed line of the cutoff (E−1max) were kept as binding peptides for a given antibody. F, an example of an epitope identified with MEME based on the binding peptides, the number, i.e., 44, of binding peptides for generating the logo is depicted on the right as “44 Peps.” The epitope was accompanied by a position-specific probability matrix from MEME. G, an example of a similarity calculation between the epitopes and the antigens. The red boxes (each consisting of six amino acids) on the antigens indicate the best-matched position-specific probability matrix (epitope) based on which the epitope–antigen similarity was calculated.