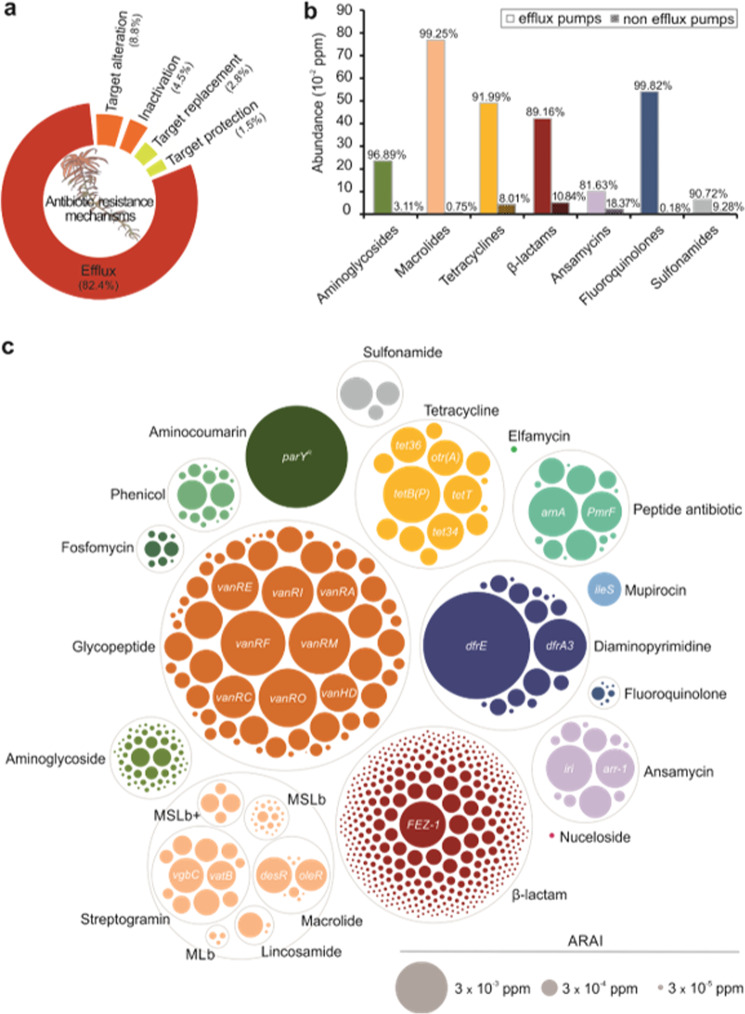
Fig. 4. The S. magellanicum metagenome comprises a highly versatile resistome.
The Illumina generated 41.8 Gbps moss metagenome from the Pirker Waldhochmoor, Austria (N46°37′38.66″, E14°26′5.66″), was aligned against the CARD sequences. a The five major resistance mechanisms presented by their relative abundance within the moss resistome. b For a selected group of antibiotic classes, the extent of efflux pump mediated and non-efflux pump mediated resistance is compared. Abundance within the metagenome is given in absolute numbers by the ARAI in ppm (≙reads per million reads), while the abundance within antibiotic classes is given as proportion in percent. c All detected non-efflux pump related ARGs grouped according to antibiotic classes. Each bubble represents one determinant with absolute abundance within the metagenome reflected by bubble size. The most abundant determinants are labelled with the gene names. MLb macrolide and lincosamide. MSLb, MLb and streptogramin. MSLb+, MSLb and oxazolidione, pleuromutilin and phenicol. CARD Comprehensive Antibiotic Resistance Database, ARAI Antibiotic Resistance Abundance Index.