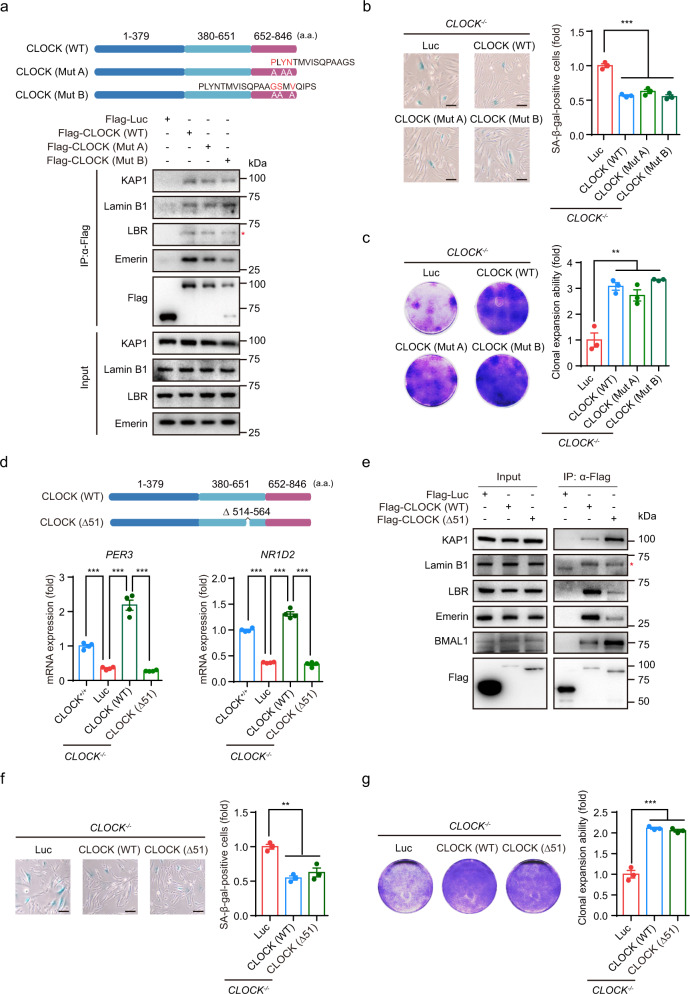
Fig. 5. CLOCK counteracts hMSC senescence independent of its transcriptional activity.
a Top, schematic diagram showing the mutation strategy for CLOCK. Bottom, Co-IP of KAP1 and nuclear lamina proteins with exogenous WT and mutant CLOCK proteins (Mut A and Mut B) in HEK293T cells. Data are representative of two independent experiments. The red asterisk indicates the LBR band pulled down by IP with Flag antibody. b SA-β-gal staining in CLOCK–/– hMSCs transduced with lentiviruses expressing Luc, CLOCK (WT), CLOCK (Mut A), or CLOCK (Mut B). The percentage of SA-β-gal-positive cells in each cell type was normalized to that in CLOCK−/− hMSCs transduced with Luc. Data are presented as means ± SEM. n = 3 biological replicates. ***P < 0.001 (two-tailed unpaired Student’s t-test). Scale bars, 100 μm. c Clonal expansion assay of CLOCK−/− hMSCs transduced with lentiviruses expressing Luc, CLOCK (WT), CLOCK (Mut A), or CLOCK (Mut B). The clonal expansion ability of each cell type was normalized to that of CLOCK–/– hMSCs transduced with Luc. Data are presented as means ± SEM. n = 3 biological replicates. **P < 0.01 (two-tailed unpaired Student’s t-test). d Top, schematic diagram showing the mutation strategy for the CLOCK Δ51 truncation. Bottom, relative mRNA levels of PER3 and NR1D2 in CLOCK+/+ hMSCs and CLOCK−/− hMSCs transduced with lentiviruses expressing Luc, CLOCK (WT), or CLOCK (Δ51). Data are presented as means ± SEM. n = 4. ***P < 0.001 (two-tailed unpaired Student’s t-test). Data are representative of two independent experiments. e Co-IP of KAP1 and nuclear lamina proteins with exogenous CLOCK (WT) and CLOCK (Δ51) proteins in HEK293T cells. Data are representative of two independent experiments. The red asterisk indicates the band of Lamin B1. f SA-β-gal staining of CLOCK−/− hMSCs transduced with lentiviruses expressing Luc, CLOCK (WT), or CLOCK (Δ51). The percentage of SA-β-gal-positive cells in each cell type was normalized to that in CLOCK–/– hMSCs transduced with Luc. Data are presented as means ± SEM. n = 3 biological replicates. **P < 0.01 (two-tailed unpaired Student’s t-test). Scale bars, 100 μm. g Clonal expansion assay of CLOCK–/– hMSCs transduced with lentiviruses expressing Luc, CLOCK (WT) and CLOCK (Δ51). The clonal expansion ability of each cell type was normalized to that of CLOCK–/– hMSCs transduced with Luc. Data are presented as means ± SEM. n = 3 biological replicates. ***P < 0.001 (two-tailed unpaired Student’s t-test).