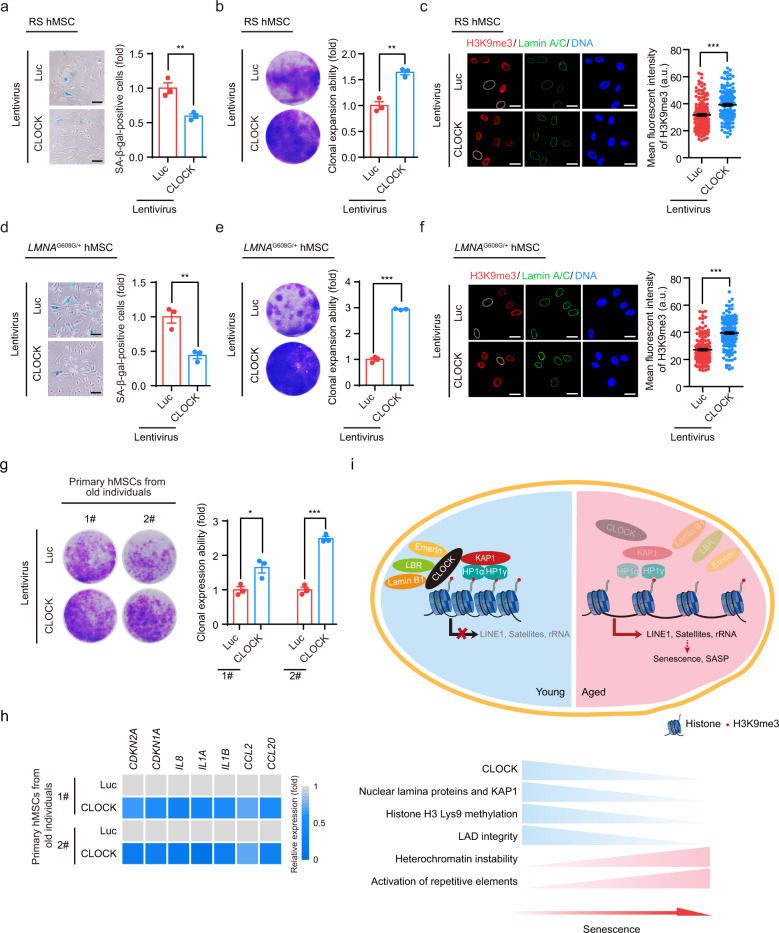
Fig. 6. CLOCK overexpression attenuates hMSC aging.
a SA-β-gal staining of replicative senescent (RS) CLOCK+/+ hMSCs transduced with lentiviruses expressing Luc or CLOCK. Data are presented as means ± SEM. n = 3 biological replicates. **P < 0.01 (two-tailed unpaired Student’s t-test). Scale bars, 100 μm. b Clonal expansion assay of RS CLOCK+/+ hMSCs transduced with lentiviruses expressing Luc or CLOCK. Data are presented as means ± SEM. n = 3 biological replicates. **P < 0.01 (two-tailed unpaired Student’s t-test). c H3K9me3 staining of RS CLOCK+/+ hMSCs transduced with lentiviruses expressing Luc or CLOCK. Dashed lines indicate the nuclear boundaries of cells with decreased H3K9me3 signals. Data are presented as means ± SEM. n = 150 cells from three biological replicates. ***P < 0.001 (two-tailed unpaired Student’s t-test). Scale bars, 25 μm. d SA-β-gal staining of HGPS-specific hMSCs transduced with lentiviruses expressing Luc or CLOCK. Data are presented as means ± SEM. n = 3 biological replicates. **P < 0.01 (two-tailed unpaired Student’s t-test). Scale bars, 100 μm. e Clonal expansion assay of HGPS-specific hMSCs transduced with lentiviruses expressing Luc or CLOCK. Data are presented as means ± SEM. n = 3 biological replicates. ***P < 0.001 (two-tailed unpaired Student’s t-test). f H3K9me3 staining of HGPS-specific hMSCs transduced with lentiviruses expressing Luc or CLOCK. Dashed lines indicate the nuclear boundaries of cells with decreased H3K9me3 signals. Data are presented as means ± SEM. n = 150 cells from three biological replicates. ***P < 0.001 (two-tailed unpaired Student’s t-test). Scale bars, 25 μm. g Clonal expansion assay of primary hMSCs that were derived from a 92-year-old individual and a 76-year-old individual and transduced with lentiviruses expressing Luc or CLOCK. Data are presented as means ± SEM. n = 3 biological replicates for two individuals. *P < 0.05; ***P < 0.001 (two-tailed unpaired Student’s t-test). h Heatmap showing the mRNA levels of the indicated genes in primary hMSCs transduced with lentiviruses expressing Luc or CLOCK. The average expression levels of the indicated genes of primary hMSCs transduced with lentiviruses expressing CLOCK were normalized to those of primary hMSCs transduced with lentiviruses expressing Luc. n = 4. i A constructive model describing the role of CLOCK in stabilizing heterochromatin during aging. In young hMSCs, CLOCK forms a complex with the heterochromatin-associated protein KAP1 and nuclear lamina proteins Lamin B1, LBR and Emerin. This complex tethers H3K9me3-enriched heterochromatin to nuclear periphery, repressing aberrant transcription of repetitive elements. In aged hMSCs, the complex is destabilized due to the downregulation of CLOCK and its binding partners, which results in heterochromatin instability and thereby transcription of repetitive elements, leading to cellular senescence and SASP.