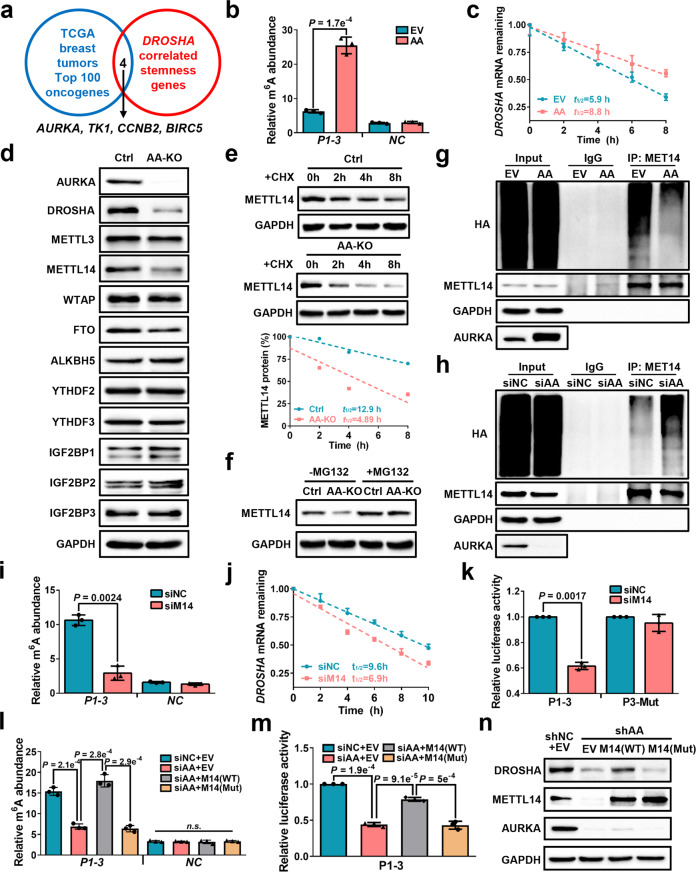
Fig. 4. AURKA-stabilized METTL14 maintains DROSHA transcript via m6A deposition.
a Comparison of DROSHA-correlated stemness genes with highly expressed oncogenes in TCGA breast tumor (top 100), common genes were listed. b meRIP-qPCR was used to determine relative DROSHA P1-3 m6A levels in AURKA-overexpressing SK-BR-3 cells. EV, empty vector, AA, overexpression of AURKA. c Stability of DROSHA mRNA in AURKA-overexpressing SK-BR-3 cells. d The expression of indicated proteins was detected in AURKA-knockout MDA-MB-231 cells by western blotting. AA-KO, knockout of AURKA. e METTL14 expression was detected in AURKA-knockout MDA-MB-231 cells treated with cycloheximide (CHX, 200 μg/mL). METTL14 proteins were quantified by densitometry and plotted as a scatter diagram at the bottom. f METTL14 expression was detected in AURKA-knockout MDA-MB-231 cells treated with MG132 (10 μM) for 8 h. g, h HEK293T cells were transfected with indicated constructs (g) or siRNAs (h) followed by co-transfection with HA-Ub and METTL14 constructs. Cells were treated with MG132 (10 μM) for 8 h before collection. The whole cell lysate was subjected to immunoprecipitation with METTL14 antibody and western blotting with anti-HA antibody to detect ubiquitylated METTL14. i meRIP-qPCR was performed to verify relative DROSHA P1-3 m6A levels in METTL14-knockdown MDA-MB-231 cells. j Stability of DROSHA mRNA in METTL14-knockdown MDA-MB-231 cells. k Relative luciferase activity of P1-3 and P3-Mut in HEK293T cells with METTL14 knockdown. l Relative DROSHA P1-3 m6A levels were detected in AURKA-deficient MDA-MB-231 cells with or without overexpression of indicated METTL14. m Relative luciferase activity of P1-3 in AURKA-deficient HEK293T cells with or without indicated overexpression of METTL14. n DROSHA, METTL14 and AURKA protein expression was identified in AURKA-deficient MDA-MB-231 cells with or without overexpression of indicated METTL14. shAA, shRNA against AURKA. Data are shown as means ± SD. P values were calculated with two-tailed unpaired Student’s t-test and P < 0.05 is considered statistically significant.