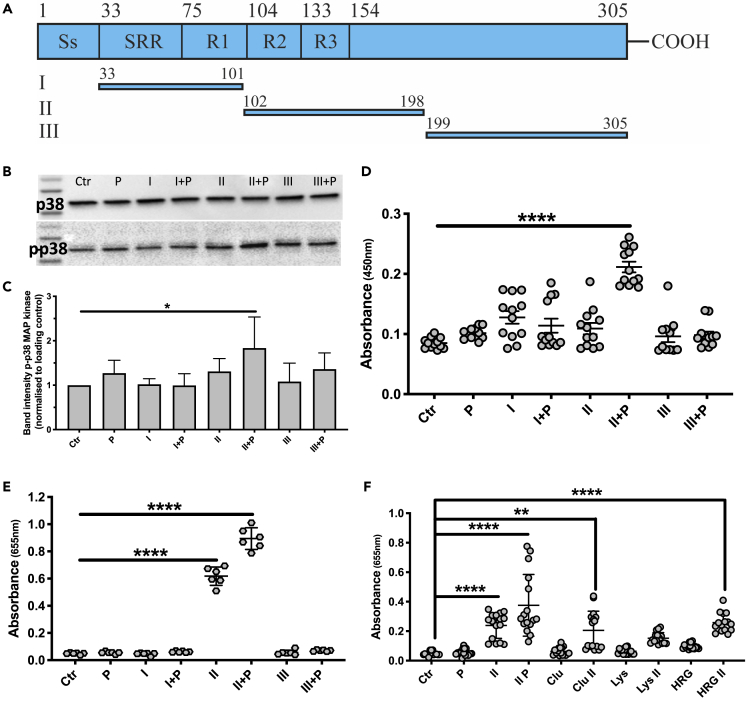
Figure 4.
Fragment II of SIC (aa 102–198) is responsible for the activation of THP1 cells
(A) Schematic structure of SIC from strain AP1 and zlocalization of the three fragments used. The signal sequence (Ss) is cleaved off during maturation. The mature secreted protein contains a short repeat region (SRR), as well as three tandem repeats (R1-R3). The numbers indicate amino acid positions, and also refer to the length of each of the SIC fragments (I-III), which are recombinantly expressed in E. coli. Based on previous publications (Fernie-King et al., 2004; Binks et al., 2005), the sizes of the fragments were chosen.
(B) Western blot analyses of THP1 cell lysates after stimulation with 5 μg/mL SIC fragments (I, II, III) +/− 2.5% plasma (P). Membranes were incubated with antibodies against p38 MAPK (p38) and phosphorylated p38 MAPK (p-p38).
(C) Quantification of phosphorylation of Western blots depicted in B. The band intensities of p-p38 samples were normalized to loading control (p38 band) and values analyzed.
(D) THP1 cells were incubated with 5 μg/mL SIC fragments +/− 2.5% plasma (P) and the release of TNFα was measured by ELISA.
(E) Activation of NF-κB by SIC fragments +/− plasma was analyzed by detecting QuantiBlue color change at 655 nm.
(F) Absorbance of QuantiBlue color change at 655 nm, as indicator of NF-κB activation after THP1 cell incubation with 5 μg/mL SIC fragment II alone and in combination with 2.5% whole plasma (P), 5 μg/mL clusterin (Clu), 5 μg/mL lysozyme (Lys) or 200 ng/mL HRG. All data represent mean ± SEM of 4-6 independent experiments, one-way ANOVA, Dunnett's multiple comparison test, with single pooled variance. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.