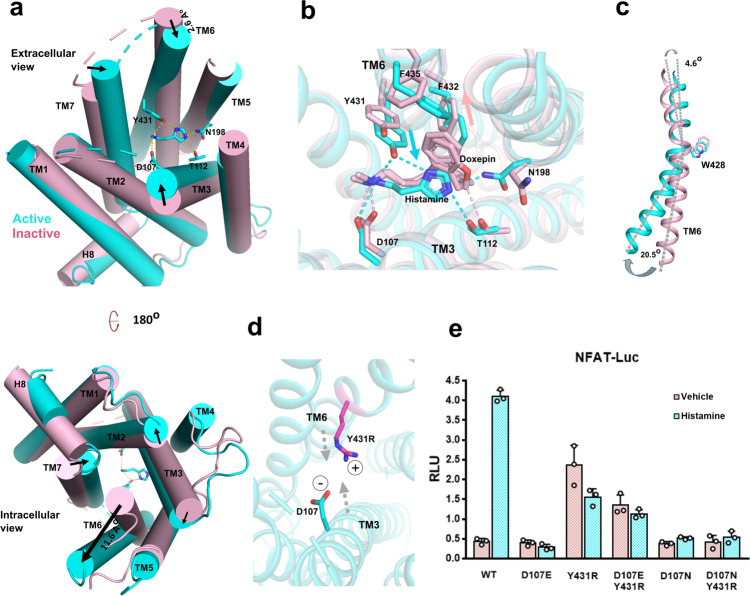
Fig. 3. Activation of H1R by the binding of histamine.
a A comparison of histamine-bound active H1R and the inverse agonist doxepin-bound inactive H1R (PDB 3RZE). The helices are shown in cylinders. Active receptor is shown in cyan and inactive receptor is shown in light pink. Up-panel is the extracellular side view and lower-panel is the intracellular side view. Arrows mark the movement of the designate part of receptor upon histamine binding. b A comparison of the agonist (histamine) binding pocket (cyan) with the inverse agonist (doxepin) binding pocket (light pink). The cyan arrow marks the movements of key residues on TM6 upon agonist (histamine) binding and the light pink arrow marks the movements of key residues on TM6 upon inverse agonist (doxepin) binding. c The movements of TM6 upon receptor activation. The W4286.48 severs as a pivot between the upside (extracelluar) movement and the downside movement of TM6. d A simple model (for illustration only) of a designed salt bridge interaction between the Y431R mutation of TM6 and with the negative charge D1073.32 of TM3. e A NFAT-RE reporter assay of the Y4316.51 and D1073.32 mutations. Histamine, 10 µM; data are presented as mean values ± SD; n = 3 independent samples. RLU, relative luciferase unit.