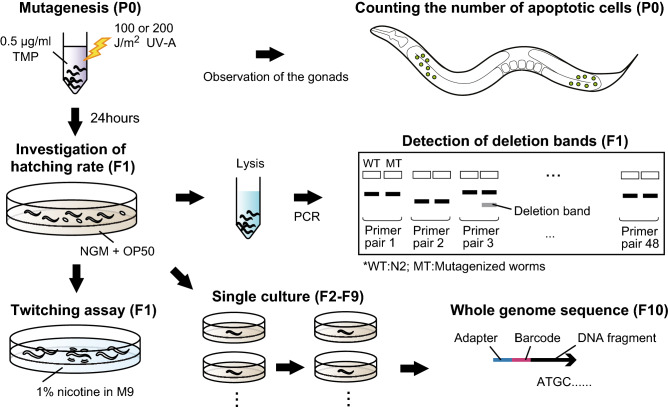
Figure 1.
Experimental design. The adult worms were exposed to TMP and irradiated with UV. Apoptotic cells in the gonads were counted 2 h after and obtained F1 eggs by washing off the adults and larvae 24 h after the mutagenesis. The hatching rates were measured the next day. The thousands of the siblings of hatched F1 larvae were soaked with 1% nicotine for the twitching assay and other thousands of F1 worms were lysed to detect deletions by PCR. For the whole genome sequencing, more than 5 F1 worms were randomly picked and cultured individually. The single culturing was repeated until the F9 generation and the starved F10 worms were used to extract genomic DNA. The extracted genome was sheared and about 120 base fragments were tagged by the adapter and barcode sequence. Finally, the mixture of the tagged DNA was sequenced using ionProton system.