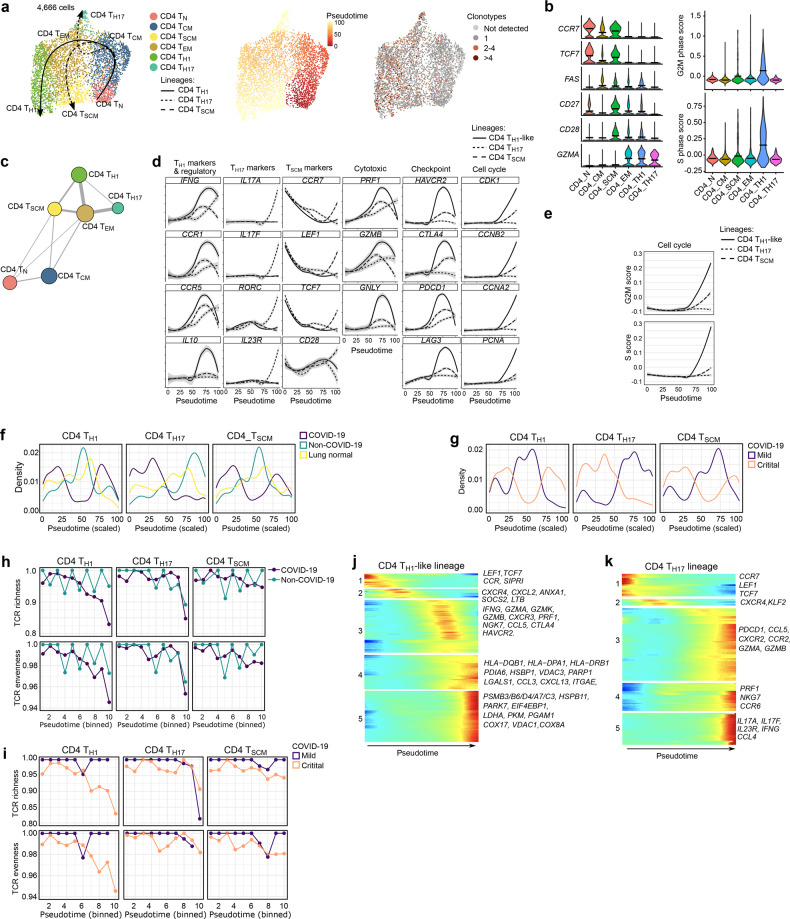
Fig. 4. CD4+ T-cell developmental trajectories in mild and critical COVID-19 BAL.
a UMAP with pseudotime trajectories based on Slingshot, showing 3 lineages (TH1-lineage, TH17-lineage and TSCM-lineage), color-coded for the CD4+ T-cell phenotypes (left), the pseudotime (middle) and the number of clonotypes (right). TSCM-cells represent a subset of minimally differentiated T-cells characterized by phenotypic and functional properties that bridge naïve and conventional memory T-cells.81 TSCM-cells include cells that show high expression of naïve (or precursor) markers (CCR7, TCF7), but not of activation markers (GZMA), high expression of memory markers (CD27, CD28 and CD95), are more proliferative compared to TN- and TCM-cells (increased G2M and S scores), increased TCR clonotype expansion compared to TN- and TCM-cells. b Naïve and memory-related marker gene expression (left), and cell cycle scoring (right) reveal additional CD4+ T-cell subclusters. TSCM-cells are characterized by naïve marker genes (CCR7, TCF7), memory markers (CD27), cell proliferation but no GZMA expression. c Analysis of clonotype sharing (thickness indicates proportion of sharing, circle size indicates clonotype counts) between the CD4+ T-cell subclusters. d Profiling of marker and functional genes along these trajectories to confirm their functional annotation. e Dynamics of T-cell proliferation along the CD4+ T-cell lineages are depicted by plotting cell cycle G2M and S scores. f Density plots reflecting the number of T-cells along the 3 CD4+ T-cell lineages stratified for non-COVID-19, COVID-19 and normal lung. g Density plots reflecting the number of T-cells along the 3 CD4+ T-cell lineages stratified for mild vs critical COVID-19. h, i TCR richness and TCR evenness along the 3 CD4+ T-cell lineages comparing non-COVID-19 vs COVID-19 (h) and mild vs critical COVID-19 (i). j, k Gene expression dynamics along the CD4+ TH1- (j) and TH17-lineage (k). Genes cluster into 5 gene sets, each of them characterized by specific expression profiles, as depicted by a selection of marker genes characteristic for each set. Differences in trajectories were assessed by Mann–Whitney test. For CD4+ TH1 and CD4+ TSCM: COVID-19 vs non-COVID-19 and lung normal (P = 1.4E-6 and 5.9E-37), For CD4+ TTh17: COVID-19 vs non-COVID-19 (P = 9.7E-12), mild vs critical COVID-19 (P = 1.3E-121).