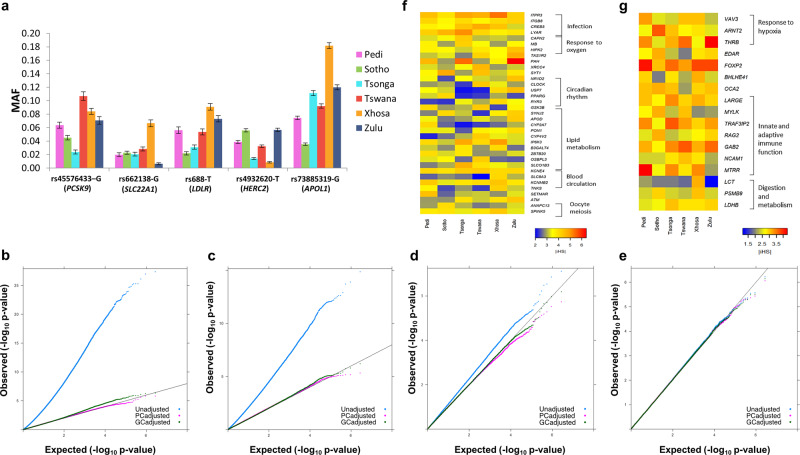
Fig. 4. Possible impact of population structure within the South-Eastern Bantu-speaking (SEB) groups on genome-wide association studies (GWASs) and evolutionary estimates.
a Allele frequency variation of some of the well-known phenotype associated SNPs. The mean and the standard error was estimated using 50 random resampling runs with 30 samples each (source data provided in Source Data file). b–e Representative QQ plots showing results from simulated-trait GWASs comparing randomly sampled participants from b Agincourt (AGT) as cases to Soweto (SWT) as controls c 62.5% AGT + 37.5% SWT participants as cases to 100% SWT participants as controls. d Random samples from SWT without Tswana as cases to random samples from SWT with Tswana as controls. e Randomly sampled individuals from SWT as cases and controls. The Observed (−log10 P-values) represent GWAS association results derived by logistic regression (two-tailed). The Expected (−log10 P-values) are those based under the null hypothesis. For b–e, blue dots represent raw P-values, whereas purple and green dots represent P-values after principal component and genomic control based correction, respectively. f Heatmap showing differences in iHS statistics for some of the SNPs that were detected as outliers (|iHS| > 4; P-value < 0.003) in at least two of the SEB groups. g Heatmap showing differences in iHS statistics for SNPs in genes previously reported to be under positive selection, that were also detected to show moderate scores in one or more of the SEB groups (|iHS| > 3, P-value < 0.05).