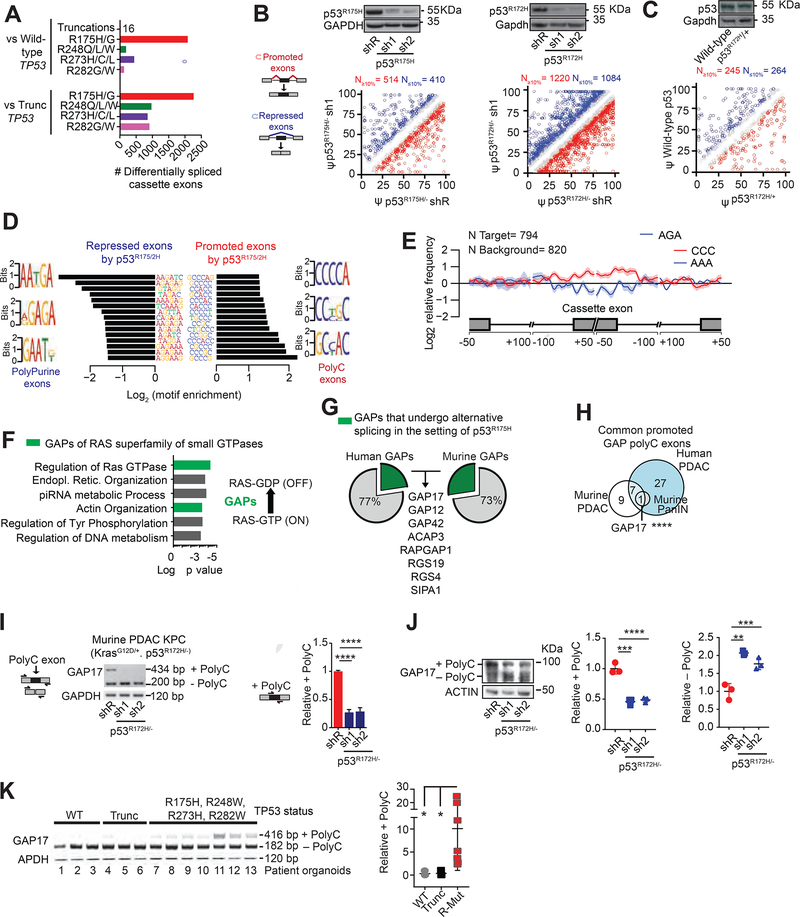
Figure 1: p53R175H promotes C-rich cassette exons, impacting expression of GAP isoforms in pancreatic cancer.
A. Differentially spliced exons in PDAC patients with missense hotspot mutations in p53 compared to those with WT TP53 and nonsense/frameshift mutations in p53 (“Trunc”). RNA-Seq from the ICGC (Bailey et al., 2016).
B-C. Differentially spliced exons whose inclusion was significantly promoted (red circles) or repressed (blue circles) in p53R175H versus control cells with knockdown of p53R175H. Splicing events quantified using ‘percent spliced in’ (PSI, or Ψ) value. Promoted and repressed exons are defined as those whose Ψ values are increased or decreased ≥10%, respectively (N). Grey circles: exons where ΔΨ is <10%. Statistical significance calculated using Kolmogorov-Smirnov test. PDAC patient organoids (KRASG12D/+; p53R172H/−) and murine KPC cells following transduction with doxycycline-inducible control (shR) or anti-p53 shRNAs (sh1 and 2) (B). RNA-seq and westerns performed after 5 days of doxycycline, n = 3. Murine PanIN organoids (KrasG12D/+; WT p53; Mist1-Cre) bearing p53R172H/+ introduced by CRISPR, compared to WT p53 (C). RNA-seq and westerns performed 20 days after confirmation of genome editing, n= 3.
D. Nucleotide motifs enriched in repressed and promoted exons by p53R175H identified from comparisons in B-C.
E. Spatial distribution and relative frequency of CCC and AAA motifs in exons promoted versus repressed by p53R172H in murine PDAC cells. Shading, 95% confidence interval by bootstrapping.
F. Left, Gene Ontology analysis of mRNAs with polyC exons based on the comparisons in B-C. p values for multiple comparisons. Green bars, biologic processes involving GAPs of RAS GTPases. Right, schema of how GAPs terminate signaling by inducing GTP hydrolysis of GTP-bound RAS.
G. Percentage of GAPs that undergo alternative splicing (AS) in the presence of p53R175H in patient and murine PDAC. GAP mRNAs with exon splicing events in both human and murine PDACs listed.
H. Intersection of GAP mRNAs that gain polyC exons by p53R175H from A-C comparisons. Fisher’s exact test p= 0.0007.
I. Qualitative and quantitative RT-PCRs of GAP17 AS in KPC cells in the presence or absence of mutant p53.
Left, schematic of primers that flank the two exons (exons 16 and 18) surrounding the polyC exon (exon 17). Upper band denotes inclusion of polyC exon, while lower band corresponds to isoform lacking the polyC exon. Right, qRT-PCR quantification of inclusion of exon 17 using primers that flank the exon junctions. Mean ± s.d, n = 3 replicates/condition. Student’s t-test.
J. Western blot and quantifications demonstrating loss of GAP17 isoforms +/− polyC exons under p53R172H knockdown or renilla control in KPC cells. Mean ± s.d, n = 3 replicates/condition. Student’s t-test.
K. RT-PCR (left) and qRT-PCRs (right) of GAP17 AS revealing distinct isoforms in patient PDAC organoids with WT p53, truncated p53 (Trunc), and hotspot mutations (using same primer approach as in (I)). Mean ± s.d, Student’s t-test.
s.d, standard deviation; n, number of repetitions. *p < 0.05, **p < 0.01, ***p < 0.001, **** p < 0.0001. See also Figures S1–S2. Tables S1–S3.