Figure 1.
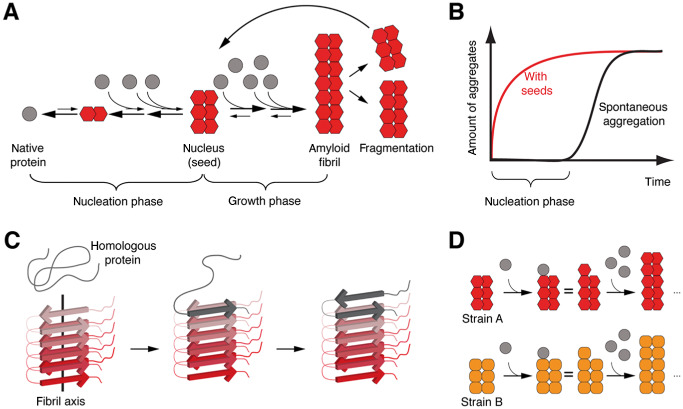
Nucleation‐dependent polymerization, seeding and templated misfolding. A. Under certain circumstances, some proteins can adopt an alternative, misfolded conformation that allows for multimerization. The initial conformational changes are kinetically unfavorable and may not occur under physiological conditions. However, once a nucleus (“seed,” putatively multimeric) is formed, the properties of the nucleus allow for relatively rapid incorporation of homologous proteins into amyloid fibrils. Fragmentation of fibrils generates more nuclei resulting in a vicious cycle of progressive protein misfolding and aggregation. B. Schematic representation of protein aggregation kinetics. Spontaneous aggregation is delayed because of the thermodynamically unfavorable nucleus formation. Addition of preformed seeds leads to immediate aggregate formation and circumvents the nucleation phase. C. Structure analysis of amyloid fibrils reveals typical sets of in‐register β‐sheets arranged parallel to the fibril axis. This structure has been suggested to serve as a template for the congruent incorporation of homologous, native or partially unfolded proteins that orient their peptide chains accordingly, hence termed templated misfolding. D. Some proteins can adopt diverse misfolded conformations that can be associated with different pathogenic properties. These conformational differences are referred to as “strains” and can be propagated by templated misfolding.
