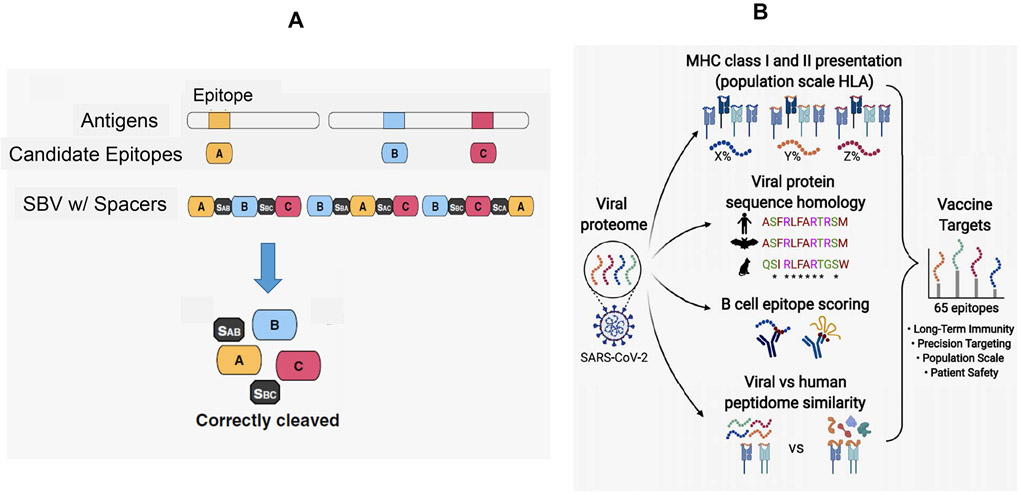
Figure 12. String-of-beads vaccines (SBVs) for multi-epitope delivery.
A. The concept of SBVs for infectious disease agents (e.g., influenza and cytomegalovirus) is based on expressing multiple epitopes from pathogen antigens separated by cleavable spacers.110 It is envisaged that the design tools for the selection of multiple epitopes and their encapsulation in suitable nanocarriers will facilitate SARS-CoV-2 epitope delivery to regional lymph nodes, where individual epitopes will be released by proteolytic processing and presented by dendritic cells. Vaccine efficiency would depend on the optimal combination of B and T-cell epitopes as well as the use of appropriate spacers to allow efficient epitope release.110 Reprinted with permission from ref 110 under the Creative Commons Attribution 4.0 International License. Copyright 2016 Springer Nature. B. Multi-epitope vaccines have received a boost from new immunoinformatic tools for vaccine design, using a series prediction tools as outlined in Table 3. In this example, Yarmarkovich et al. report the design of multi-epitope vaccines that deliver 65 x 33-mer SARS-CoV-2 peptides, making use of the following guidelines: (i) selection of peptide sequences from 15 related coronaviruses; (ii) epitope ability to activate CD4+ and CD8+ T-cells by enabling interactions with diverse HLA gene sequences; (iii) B-cell activation by linear and conformational epitope sequences; (iv) high immunogenicity through sequence selection that is significantly dissimilar to the self-proteome; (v) vaccine safety.109 Reprinted with permission from ref 109 under the Creative Commons License CC-BY 4.0. Copyright 2020 The Authors.