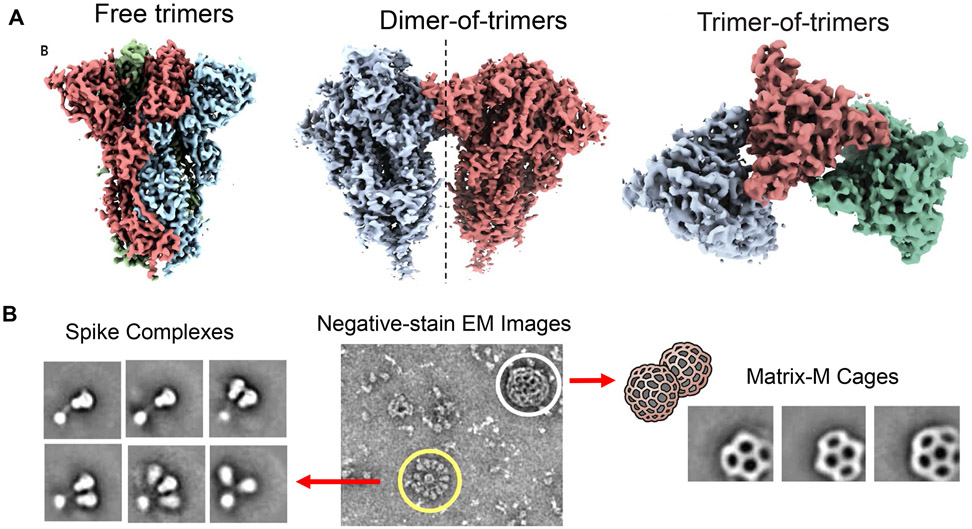
Figure 8. COVID-19 vaccine development through self-assembly of stabilized, full-length SARS-CoV-2 S subunits into nanoparticles.
A. Novavax developed a full-length S protein subunit vaccine that is expressed as a stabilized protein with mutational deletion of the furin cleavage site plus 2 proline (2-P) substitutions (K986P and V987P) that confer an "RBD-up" conformation.61 This leads to expression of SARS-CoV-2 3Q-2P spike protein as a pre-fusion complex. When reconstituted in Polysorbate 80 (PS 80), the protein adopts tertiary structures that include free trimers, dimers-of-trimers, trimers-of-trimers or multi-trimer rosettes (with as many as 14 trimer transmembrane domains being enclosed in micellar PS 80 cores).61 Moreover, the vaccine is further reconstituted with the Matrix-M adjuvant. Reprinted with permission from ref 61. Copyright 2020 AAAS. B. Negative stain electron microscopy of the full-length spike (reconstituted in PS 80), admixed with the cage-like Matrix-M component (from plant origin).61,209 The spike rosettes are circled in yellow and Matrix-M adjuvant cages are circled in white.90 The imaging confirms the presence of trimeric spike as free trimers or as multitrimer rosettes. Matrix-M does not appear to interact with the spike nanoparticles. Reprinted with permission from ref 209. Copyright 2020 AAAS.