Abstract
It has become evident that the actions of pro-inflammatory cytokines and/or the development of a cytokine storm are responsible for the occurrence of severe COVID-19 during SARS-CoV-2 infection. Although immunomodulatory mechanisms vary among viruses, the activation of multiple TLRs that occurs primarily through the recruitment of adapter proteins such as MyD88 and TRIF contributes to the induction of a cytokine storm. Based on this, controlling the robust production of pro-inflammatory cytokines by macrophages may be applicable as a cellular approach to investigate potential cytokine-targeted therapies against COVID-19. In the current study, we utilized TLR2/MyD88 and TLR3/TRIF co-activated macrophages and evaluated the anti-cytokine storm effect of the traditional Chinese medicine (TCM) formula Babaodan (BBD). An RNA-seq-based transcriptomic approach was used to determine the molecular mode of action. Additionally, we evaluated the anti-inflammatory activity of BBD in vivo using a mouse model of post-viral bacterial infection-induced pneumonia and seven severely ill COVID-19 patients. Our study reveals the protective role of BBD against excessive immune responses in macrophages, where the underlying mechanisms involve the inhibition of the NF-κB and MAPK signaling pathways. In vivo, BBD significantly inhibited the release of IL-6, thus resulting in increased survival rates in mice. Based on limited data, we demonstrated that severely ill COVID-19 patients benefited from BBD treatment due to a reduction in the overproduction of IL-6. In conclusion, our study indicated that BBD controls excessive immune responses and may thus represent a cytokine-targeted agent that could be considered to treating COVID-19.
Abbreviations: DMEM, Dulbecco’s modified Eagle’s medium; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; Pam3CSK4, Pam3-Cys-Ser-Lys-Lys-Lys-Lys; PolyI:C, polyinosinic-polycytidylic acid; NF-κB, nuclear factor kappa-B; MAPK, mitogen-activated protein kinase; ISRE, interferon-sensitive response element; IRF, interferon regulatory factor; PaO2, arterial partial pressure of oxygen; FiO2, fraction of inspired oxygen; CRP, C-reactive protein; CT, computed tomography
Keywords: COVID-19, Cytokine storm, Inflammation, Traditional Chinese medicine, Babaodan
Graphical Abstract
1. Introduction
There is a rapidly evolving situation regarding coronavirus disease 2019 (COVID-19) that is rapidly spreading from person to person and is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since the initial outbreak in Wuhan, China, COVID-19 has officially become a pandemic. To date, reports on confirmed cases of COVID-19 and the associated deaths continue to accumulate globally (https://who.sprinklr.com). Consequently, numerous efforts are currently underway to develop therapeutic agents and vaccines [1].
It has become evident that robust pro-inflammatory cytokine production, also known as the cytokine storm, is indicative of severe COVID-19 and may lead to pulmonary pathology or multiple organ failure that is frequently observed in patients. As a result, substantially elevated serum levels of pro-inflammatory cytokines, including interleukin (IL)−6, IL-1β, and tumor necrosis factor (TNF)-α, were detected in the majority of patients with severe COVID-19. Specifically, the presence of IL-6 is associated with a relatively poor outcome in patients with severe COVID-19, pneumonia, or acute lung injury [2], [3], [4], [5], [6]. Therefore, a cytokine-targeted therapeutic strategy has been proposed for the treatment of patients with severe COVID-19 [7], [8].
Inflammatory monocytes are critical for controlling the profile and the length of production of pro-inflammatory cytokines [9]. This is also true for COVID-19 infections. Zhang et al. demonstrated that inflammation-related phenotypic alterations in peripheral blood monocytes could be observed, the majority of which were characterized by the production of IL-6, IL-10, and TNF-α, the extent of which was correlated with patient outcome [10]. Accordingly, a single cell analysis-based transcriptional approach revealed that an increase in IL-1β-producing monocytes could serve as an indicator for the early recovery stages in COVID-19 patients [11]. It has been recognized that during the course of COVID-19, SARS-CoV-2 enters airway epithelial cells via angiotensin-converting enzyme 2 (ACE-2). Concurrently, macrophages also release large amounts of cytokines to initiate inflammatory responses, where the severity is likely related to different immune factors rather than to receptor binding [12]. In this scenario, a selective Bruton tyrosine kinase inhibitor (Acalabrutinib) was tested in 19 patients that were hospitalized with severe COVID-19 in a prospective off-label clinical study. The results revealed that this treatment improved oxygenation in the majority of patients by ameliorating inflammatory processes such as those mediated by C-reactive protein (CRP) and IL-6 [13]. Therefore, treatment that mitigates the excessive inflammation induced by the actions of macrophages may be beneficial for controlling the inflammatory status of patients with severe COVID-19.
A typical pathogen presents a combination of toll-like receptor (TLR) ligands during infection. TLRs share the common adapter proteins myeloid differentiation factor 88 (MyD88) and TIR domain-containing adapter inducing IFN-β (TRIF) to initiate downstream signaling pathways [14], [15], [16]. In contrast to individual TLR activation, multi-TLR activation may lead to a highly synergistic cytokine response in macrophages [17], ultimately leading to the production of late-peaking T cell-polarizing cytokines such as IL-6 [18]. This phenomenon also appears to be true for COVID-19. For example, SARS-CoV-2 binds to TLRs to stimulate the induction of IL-1β and IL-6, and this eventually leads to lung inflammation that may progress to fibrosis [19]. In this regard, although no exact cell models are available to match the pathology of COVID-19, MyD88 and TRIF co-activation in macrophages could be applied as a cell model for examining cytokine-targeted treatment regimens.
Based on traditional uses and clinical practices, it is evident that traditional Chinese medicine (TCM) possesses therapeutic potential. For example, recently published data demonstrated that lianhuaqinwen exhibited anti-viral and anti-pro-inflammatory cytokine production activities [20]. Specific to cytokine-targeted effect, TCMs within the “heat-clearing and detoxifying (qingre jiedu)” category have been shown to display strong anti-inflammatory activities that include satisfactory clinical therapeutic effects against acute lung injury [21], [22]. Babaodan (BBD) is a TCM formula composed of Bovis Calculus (Niuhuang), Fel Serpentis (She dan), Cornu Saigae Tataricae (Lingyangjiao), Margarita (Zhenzhu), Moschus (Shexiang), and Notogiseng Radix et Rhizoma (Sanqi). According to TCM theory, BBD treatment induces effects that include “clearing and humid heat (qingli shire)”, “activating blood and removing toxicity (huoxue jiedu)”, and “curing jaundice and relieving pain (quhuang zhitong)”. In clinical practice, BBD has been widely used for the treatment of infectious diseases, including viral hepatitis and non-infectious liver injury [23], [24], [25], [26], [27]. Due to its promising anti-inflammatory activities, we hypothesized that BBD could be a potential candidate for anti-cytokine storm-based therapies.
In our current study, we determined the effect of BBD on inflammatory conditions and explored the molecular mode of action of this compound. We used Pam3CSK4 (TLR1/2 agonist) and Poly(I:C) (TLR3 agonist) as dual stimulation agents on RAW 264.7 macrophages to co-activate MyD88- and TRIF-dependent signaling pathways that could satisfactorily stimulate the development of a cytokine storm. In addition to the evaluation of the production of pro-inflammatory cytokines through the use of RT-qPCR, RNA sequencing (RNA-seq) was used for transcriptional profiling analysis. Additionally, we utilized the influenza virus PR8 and Staphylococcus aureus (SA) consequence infection model that could induce significant inflammatory lung injury in mice, and we applied BBD prior to SA infection. Survival rates, pathological morphology of lung tissues, and cytokine levels of TNF-α and IL-6 in bronchoalveolar lavage fluid (BAFL) were evaluated. Eventually, we analyzed whether BBD could be beneficial for the treatment of severe COVID-19; a clinical evaluation of the off-label use of BBD was carried out for seven eligible patients.
2. Material and methods
2.1. BBD preparation
Babaodan capsules (BBD, Med-drug permit no. Z10940006) were obtained from Xiamen Traditional Chinese Medicine Co., Ltd., Shanghai Pharma (Xiamen, China). In the current study, for mice experiments a BBD (Batch no. 20170205) suspension was prepared with distilled water and intragastrically (i.g.) administered at a dose of 50 μL (1 g/kg bw). This dosage is designed as three-fold the regulator-administrated amount of BBD for humans (1.8 g/day). For cell stimulation experiments, BBD (Batch no. 20170205) solution was prepared according to published protocols with modifications [28]. Briefly, BBD was resuspended in cell culture medium, sonicated on ice for 30 min, and passed through a 0.22 μM filter to prepare the 2 mg/mL stock solution. For patient treatment, BBD (Batch no. 20190906) was prescribed at a dosage of 3 capsules, p.o., bid (1.8 g/day) for 14 days.
2.2. Cells
Mouse macrophage RAW264.7 cells were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin and streptomycin at 37 °C with 5% CO2. A total of 3 × 105 cells were grown in 6-well plates and were pretreated with BBD solution or medium for 24 h prior to treatment with poly(I:C), Pam3CSK4 (both from InvivoGen, San Diego, CA, USA), or a combination of both. At 8 h post stimulation, the cells were collected for RT-qPCR or RNA-seq. At 24 h post stimulation, cell supernatants were collected for western blotting and ELISA. At the indicated times post stimulation, cells were harvested for protein extraction and western blotting.
2.3. Microorganisms
Stains of influenza virus PR8 (A/PR/8/34, H1N1) and S. aureus were maintained in our laboratory. PR8 was grown in Madin-Darby canine kidney (MDCK) cells as previously described [29]. A single stock of the virus was used in this study. Bacteria were grown in tryptic soy broth medium and quantified according to the OD600-based bacterial growth curve and colony forming unit (CFU) assays [30], [31].
2.4. Animals
The 6–8 week-old female C57BL/6 mice weighing –16–21 g were purchased from Shanghai Laboratory Animal Company (SLAC, Shanghai, China). The mice were housed in rooms with a 12-hour light/dark cycle and were provided with free access to food and water. The animal care and experimental protocols were approved by the Animal Care and Use Committee of Zhejiang University.
The mice were divided into three groups that included (1) the control group that was treated with PBS, (2) the model group (PR8+SA) that was co-infected with PR84 and SA, and (3) the BBD treatment group (PR8+SA+BBD) that was intragastrically administered a single dose of BBD (1 g/kg body weight [bw]) at 2 h prior to SA infection.
Post-influenza S. aureus infection was established as previously reported [32], [33]. Briefly, after anesthesia with 1.5% isoflurane on day 0, the mice were infected intranasally with 200 plaque-forming units (PFU) of PR8 virus. Five days (Day 5) after influenza infection, the mice were infected with 5 × 107 or 2.5 × 107 CFU of SA or PBS (50 μL/mouse) by intratracheal instillation.
Of these, one set of experiments was performed to assess the survival rates of the mice (n = 16 for model group, n = 15 for BBD treatment group, i.t., 5 × 107 CFU of SA) by examining them every 4 h for up to 3 days. For the other experiments, SA was administered at a dosage of 2.5 × 107 CFU of SA (n = 3–4 per group). At 2, 6, 12, and 24 h post SA infection, bronchoalveolar lavage fluid (BALF) was collected for ELISA. At 12 and 24 h post SA infection, lung homogenates were collected for colony forming unit (CFU) assay analyses, and at 24 h post SA infection, whole lungs were extracted for hematoxylin and eosin (H&E) staining (Servicebio, Hangzhou, China).
2.5. Western blotting
Cells were lysed in 1% NP40 (Beyotime Biotechnology, China) containing 1 mM phenylmethylsulfonylfluoride (PMSF) and a protease inhibitor cocktail (CST, Danvers, MA, USA). Total proteins (40 μg) were separated by SDS-PAGE and blotted onto polyvinylidene fluoride (PVDF) membranes. The membranes were probed with antibodies against phospho-NF-κB p65 (Ser536), NF-κB p65, Phospho-p44/42 MAPK (Erk1/2), p44/42 MAPK (Erk1/2), phospho-p38 MAPK (Thr180/Tyr182), p38 MAPK, phospho-SAPK/JNK (Thr183/Tyr185), and GAPDH (all from CST) followed by exposure to horseradish peroxidase-conjugated secondary antibodies (Lianke, Hangzhou, China, http://www.liankebio.com/). ECL reagent was used to develop the membrane, and the signals were detected using a ChemiDoc™ Touch Imaging System (Bio-Rad, Richmond, CA, USA).
2.6. ELISA
Cell-free BAL fluid was used for enzyme-linked immunosorbent assays (ELISA) to measure the concentration of tumor necrosis factor (TNF)-α and interleukin (IL)−6 (kits from eBioscience, ThermoFisher, USA) according to the manufacturer’s protocol. The online software elisaanalysis.com (http://elisaanalysis.com/app) was used to calculate cytokine concentrations, where a 4-parameter logistic non-linear regression analysis of the standard curve was selected.
2.7. Quantitative reverse transcription-PCR (qRT-PCR)
Total RNA was extracted using the TRIzol reagent (Cwbio, Beijing, China). cDNA was synthesized using a HiFiScript cDNA synthesis kit (Cwbio). One microgram of total RNA and random primers were used for the reaction. Real-time semiquantitative PCR was performed in triplicate using the SYBR Green PCR assay and a CFX-Touch PCR System (Bio-Rad). The primer sequences are listed as follows. β-actin: forward primer: 5′-GTA TCC TGA CCC TGA AGT ACC-3′, reverse primer: 5′-TGA AGG TCT CAA ACA TGA TCT-3′; TNF-α: forward primer: 5′-AAT AAC GCT GAT TTG GTG A-3′, reverse primer: 5′-ACC CGT AGG GCG ATT ACA-3′; IL-6: forward primer: 5′-TTC CAG AAA CCG CTA TGA-3′, reverse primer: 5′-GGT TGT CAC CAG CAT CAG-3′. β-Actin was amplified as an endogenous reference gene. The relative expression level was calculated according to the ΔΔCt method using Bio-Rad CFX Manager (Bio-Rad) and expressed as fold change relative to the control.
2.8. RNA-sequencing and data analysis
RNA samples were sent to the Beijing Genomic Institution (BGI, Shenzhen, China) for mRNA preparation and RNA-seq analyses. Library construction and sequencing were performed using a BGISEQ-500. Clean tags were mapped to the reference genome using HISAT2 [34]. The original sequencing data were deposited in the public database of the NCBI BioProject under the project ID PRJNA578173.
Gene expression was calculated according to the fragments per kilobase of transcript per million mapped reads (FPKM) method using an RESM quantification tool [35] and is presented as the mean FPKM. Differentially expressed genes (DEGs) between the two groups were defined according to the absolute log 2 (fold change) > 2 and false discovery rate (adjusted P value) < 0.001. A Venn diagram was constructed to obtain co-expressed DEGs between or among samples using the R package VennDiagram (Version 1.6.20). KEGG pathway enrichment analysis was performed using the clusterProfiler package [36]. Transcription factor (TF) enrichment analysis was performed using the online analysis tool DiRE (https://dire.dcode.org/) referring to the mouse genome (mm9), a random set of 5000 genes as the background genes, and UTR ECRs plus promoter ECRs as target elements [37]. Occurrence and TF importance were assigned to the obtained TFs. The top 10 TFs were selected as the primary TFs obtained from enrichment.
2.9. Clinical observation
Seven patients with severe COVID-19 were eligible for BBD treatment. All 7 patients were treated with Babaodan at a dosage of 3 capsules, p.o., bid (1.8 g/day) for 14 days. The study was conducted at the Department of Respiratory and Critical Medicine, Taizhou Hospital of Zhejiang Province, Wenzhou Medical University, Taizhou, Zhejiang, China. Clinical outcomes were compared before and after BBD treatment. Written informed consent was obtained from all patients. This study was approved by the ethics committee of the hospital. Changes in clinical presentations, Pao2/Fio2, serum inflammatory cytokine IL-6, CRP, and lung CT scan results were recorded prior to treatment and after 2, 7, and 14-days of BBD application.
2.10. Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM). Significant differences between groups were detected according to multi-factor analysis of variance (one-way ANOVA) and t-test. Survival curves were compared using the log-rank (Mantel-Cox) test. Unless otherwise specified, all statistical analyses were conducted using GraphPad Prism (version 6.0; La Jolla, CA, USA). Statistical significance was set at P < 0.05.
3. Results
3.1. BBD inhibits robust production of pro-inflammatory cytokines in Raw 264.7 macrophages
First, we utilized TLR1/2-MyD88 and TLR3-TRIF dual-activation macrophage cell models to investigate the anti-inflammatory effect of BBD. Additionally, an RNA-seq-based transcriptomic approach was used to explore the key signaling pathways responsible for the actions of BBD.
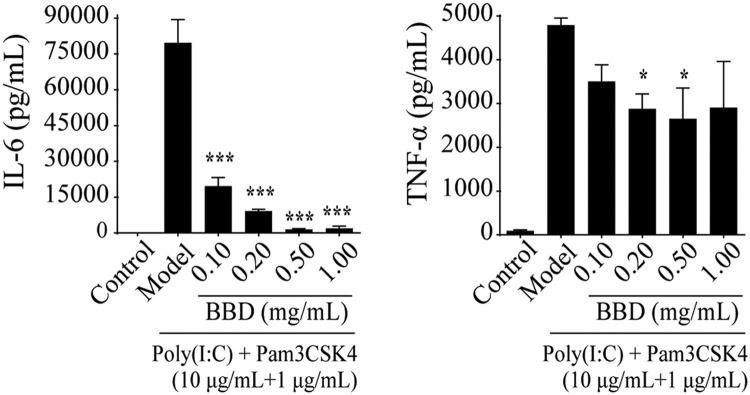
To achieve this, we employed a combination treatment using Pam3CSK4 (10 μg/mL) and Poly(I:C) (1 μg/mL) to stimulate RAW264.7 cells for 24. As expected, abundant amounts of IL-6 and TNF-α were detected. Pre-treatment of the cells with BBD for 24 h abolished the production of both of these cytokines in a dose-dependent manner ( Fig. 1). The inhibitory effect was not due to cell toxicity resulting from BBD treatment according to the results of a cell viability assay.
Fig. 1.
Babaodan (BBD) inhibits TLR1/2 and TLR3 dual stimulation-induced cytokine expression in Raw 264.7 macrophages. Raw 264.7 cells were grown on 12-well plates and incubated with different doses of BBD for 24 h, and this was followed by treatment with a combination of Poly(I:C) (10 μg/mL) and Pam3CSK4 (100 ng/mL) for 24 h. Supernatants were collected, and TNF-α and IL-6 concentrations were analyzed using the corresponding ELISA kits. *P < 0.05, **P < 0.01,***P < 0.001 vs. model group.
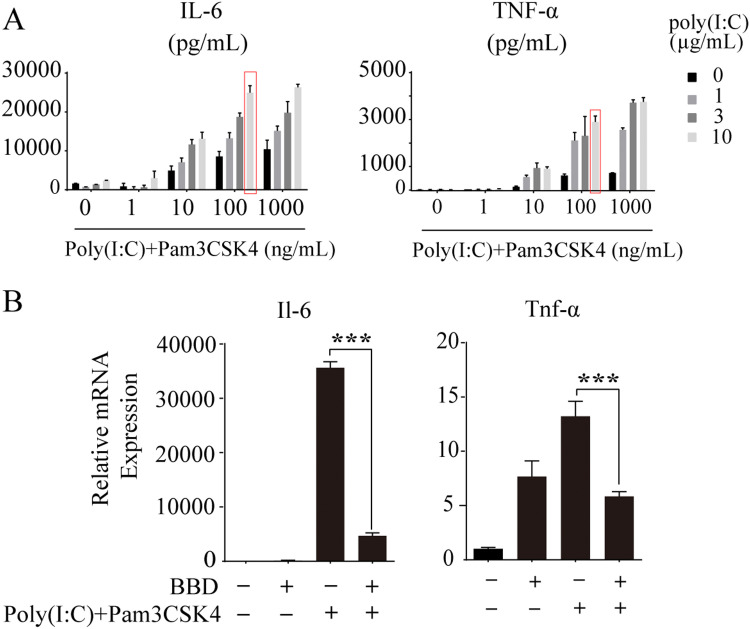
We also optimized the concentrations of Pam3CSK4 and Poly(I:C) to maximize the effect of the MyD88- and TRIF-based dual activation strategies. Raw 264.7 cells were stimulated with Poly(I:C) at 1, 3, and 10 μg/mL or Pam3CSK4 at 1, 10, 100, and 1000 ng/mL or with a combination of both stimuli. Twenty-four hours after stimulation, the production of IL-6 and TNF-α was measured. As indicated in Fig. 2A, a combination of 10 μg/mL and 100 ng/mL of Poly(I:C) and Pam3CSK4, respectively, elicited the maximum effect with regard to both IL-6 and TNF-α production. Based on this, these concentrations were used in subsequent experiments. Under these experimental conditions, BBD still exhibited powerful anti-inflammatory effects according to the results of RT-qPCR analyses (Fig. 2B).
Fig. 2.
BBD abolishes IL-6 and TNF-α production after optimized TLR1/2 and TLR3 dual activation of Raw264.7 macrophages. (A) Raw 264.7 cells were grown on 12-well plates and incubated with different combinations of Poly(I:C) and Pam3CSK4 for 24 h. Supernatants were collected, and TNF-α and IL-6 concentrations were determined using the corresponding ELISA kits. (B) Raw 264.7 cells were grown on 6-well plates and incubated with 1 mg/mL of BBD for 24 h, and this was followed by treatment with a combination of Poly(I:C) (10 μg/mL) and Pam3CSK4 (100 ng/mL) for 8 h. IL-6 and TNF-α mRNA levels were detected by RT-qPCR. *** P < 0.001 vs. model group.
3.2. Analysis of RNA-seq data
A detailed summary of the sequencing data for each sample is provided in Supplementary Table S1. A total of 19,178, 18,510, and 19,174 genes were used for subsequent analyses of the control, model, and BBD groups, respectively.
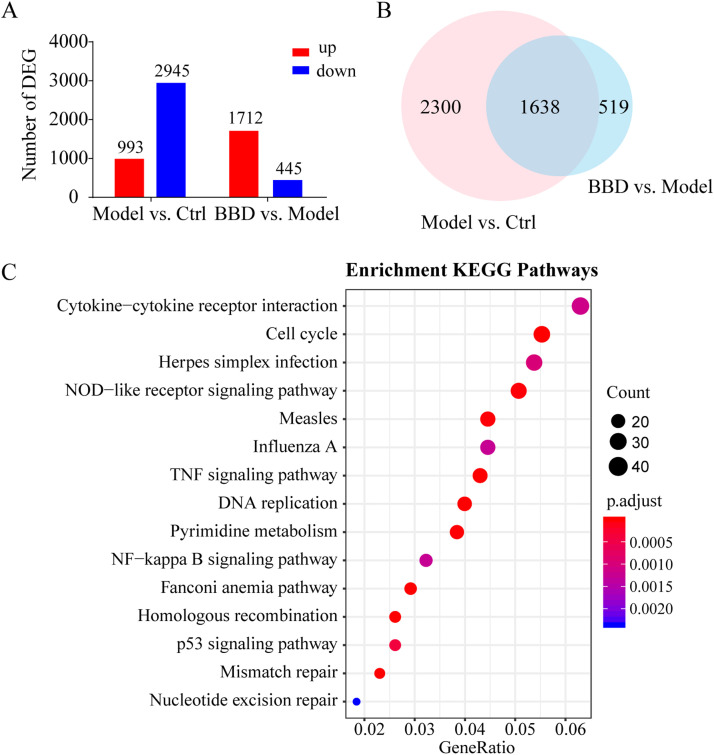
In comparison to the control group, 993 genes were upregulated in the model group and 2945 genes were downregulated (Fig. 4A and Supplementary Table S2, control vs. model). Compared to the model group, 445 genes were upregulated and 1712 were downregulated in the BBD treatment group ( Fig. 3A and Supplementary Table S2, model vs. BBD).
Fig. 3.
Gene expression profiling by RNA-seq. Raw 264.7 cells grown on 6-well plates were treated with a combination of 10 μg/mL Poly(I:C) and 100 ng/mL Pam3CSK4 (model) for 8 h with or without pre-treatment with 1 mg/mL of Babaodan (BBD) for 24 h. RNA samples were collected for RNA-seq analysis. Two samples were analyzed per group. Differentially expressed genes (DEG) are identified by absolute log2 (FC) > 2 and P < 0.05 values. (A) Number of up- and downregulated genes in the model and treatment groups. (B) Venn diagram summary of DEGs between the control vs. model and model vs. BBD groups. (C) Top 15 KEGG pathway enrichment scores of co-regulated DEGs in the model vs. control and BBD vs. model groups.
As shown in the Venn diagram, among the 3938 differentially expressed genes (DEGs) affected by TLR1/2 and TLR3 co-activation, 1638 DEGs were detected in response to BBD treatment (Fig. 3B and Supplementary Table S3). KEGG enrichment analysis was performed on 1638 co-regulated DEGs in the model vs. control and BBD vs. model groups. According to the annotation results, the top 15 KEGG pathway enrichment scores were classified as related to cytokine-cytokine receptor interaction, the cell cycle, herpes simplex virus infection, NOD-like receptor signaling, measles and influenza A virus infections, TNF signaling, DNA replication, pyrimidine metabolism, and NF-κB signaling (Fig. 3C and Supplementary Table S4).
3.3. Differential expression of pro-inflammatory cytokine genes following BBD treatment
In our analysis, we focused on the genes encoding inflammatory cytokines. Based on the KEGG gene database and the gene expression profile from our current study, a list of 311 cytokine-encoding genes and interferon-stimulated genes (ISGs) was selected for further analyses (Supplementary Table S5). The top 20 inflammatory DEGs are listed in Table 1. In accordance with the results from qPCR and ELISA assays, IL-6 is among the TOP20 regulated cytokines that occurred in models vs. control and BBD vs. model groups (Table 1). BBD could also reduce the expression of TNF by approximately 3-fold, and this represented a significant reduction, as TNF was upregulated by as much as 39-fold in response to the stimulation (Table S2).
Table 1.
Top 20 inflammatory DEG regulated by BBD in comparison to the model group.
| Symbol | Fold change (log2, model vs. control) | Fold change (log2, BBD vs. model) |
|---|---|---|
| Ccl20 | 6.84 | -5.88 |
| Ifnb1 | 6.63 | -4.67 |
| Ccl17 | 11.10 | -4.28 |
| Nr4a3 | 2.61 | -4.20 |
| Lif | 7.21 | -4.16 |
| Tslp | 4.55 | -3.90 |
| Ccl5 | 10.24 | -3.89 |
| Il12b | 7.09 | -3.80 |
| Il1b | 11.44 | -3.69 |
| Gbp5 | 5.83 | -3.62 |
| Pdgfrb | 1.41 | -3.61 |
| Ccrl2 | 4.39 | -3.59 |
| Ccl22 | 10.40 | -3.58 |
| Gbp2 | 7.08 | -3.55 |
| Parp14 | 4.00 | -3.32 |
| Usp18 | 6.57 | -3.27 |
| Cd40 | 5.41 | -3.23 |
| Dhx58 | 4.25 | -3.16 |
| Il6 | 14.17 | -3.14 |
| Ifi44 | 4.11 | -3.12 |
We also assessed the expression of macrophage activation markers such as IL-10 and of genes related to the production of iNOS or reactive oxidative species. With the exception of NOS2, Pam3CSK4 and Poly(I:C) stimulation did not increase the expression of NOS3, NOS1, and IL-10, and BBD exerted no or limited regulatory effects on these genes (Table S2).
3.4. Transcription factor analysis of inflammatory DEG following BBD treatment
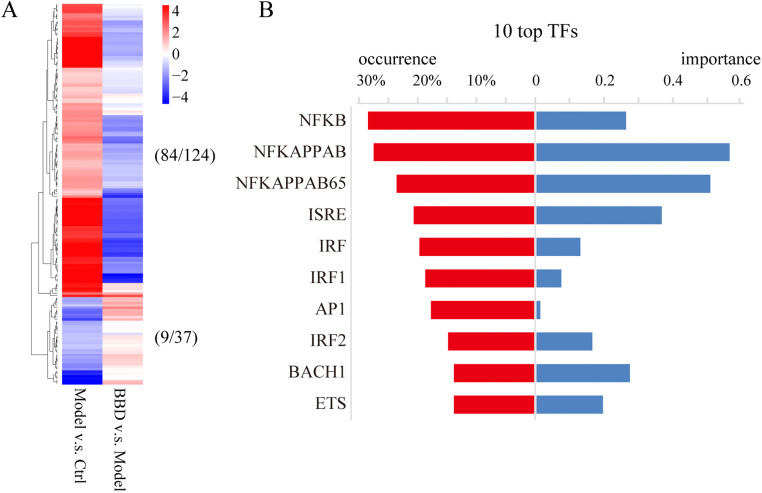
For inflammatory cytokine genes, a total of 124 genes were upregulated in the model group compared to levels in the control group, while 37 genes were downregulated. Among these genes, BBD was determined to upregulate and downregulate the expression of 84 and 9 genes, respectively, in comparison to levels in the control group ( Fig. 4A and Supplementary Table S6). Transcription factor enrichment analysis was performed on the 84 downregulated inflammatory DEGs (BBD vs. model, Fig. 4A and Supplementary Table S6), and the top 10 transcription factors with the highest occurrence are listed in Fig. 4B. Among them, NF-κB, AP-1, ISRE, and IRF were identified.
Fig. 4.
Transcriptional profiling changes in inflammatory DEG result from BBD treatment. (A) Heatmap of relative gene expression (log2[FC]) for the 124 inflammatory genes that were upregulated and the 37 inflammatory genes that were downregulated in the model vs. control groups. BBD regulates the expression of 84 and 9 of these categories of genes, respectively, compared to the control group. Blue, downregulation; red, upregulation. (B) Transcription factor (TF) enrichment analysis was performed for the 84 downregulated inflammatory DEGs, and the top 10 TFs with the highest importance are listed.
3.5. The anti-inflammatory effect of BBD is related to the NF-κB and MAPK signaling pathways
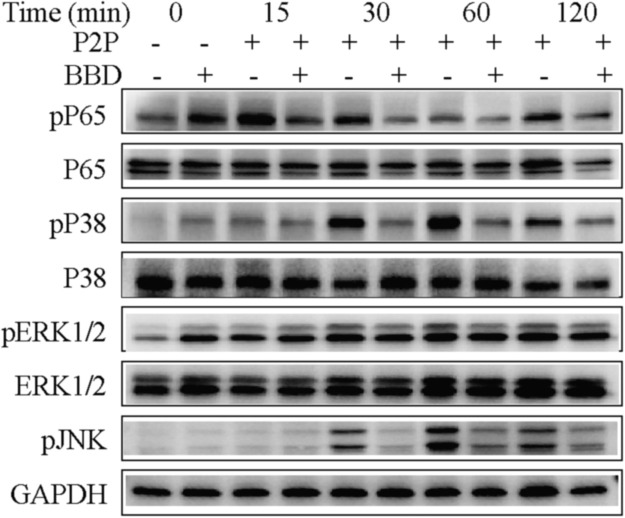
Based on the transcription factor enrichment analysis results, we investigated the activation of relevant intracellular NF-κB and MAPK (JNK, p38, and ERK) signaling pathways triggered by TLR1/2 and TLR3 co-stimulation, and we also investigated the influence of BBD pre-treatment in this context. To this end, Raw264.7 cells were stimulated with a combination of Poly(I:C) at 10 μg/mL and Pam3CSK4 at 100 ng/mL for 15, 30, 60, and 120 min, respectively, and then subjected to western blot analysis to examine the phosphorylation status of NF-κB p65, p38, JNK, and Erk1/2. As shown in Fig. 5, when the same amount of total protein was used, treatment with Poly(I:C) and Pam3CSK4 increased the phosphorylation levels of NF-κB p65, p38, and JNK, and this was inhibited by BBD.
Fig. 5.
BBD inhibits NF-κB and MAPK signaling in macrophages promoting excessive inflammatory processes. Raw 264.7 cells were grown on 6-well plates and incubated with 1 mg/mL BBD for 24 h, and this was followed by treatment with a combination of Poly(I:C) (10 μg/mL) and Pam3CSK4 (100 ng/mL) (P2P) for 15, 30, 60, and 120 min. The cells were harvested at the indicated time points after stimulation. Cell lysates were resolved by SDS-PAGE, blotted, and analyzed using the respective antibodies. One representative experiment of three is shown.
3.6. BBD exerts a pronounced anti-inflammatory effect in mice
Next, we investigated if BBD could mitigate the inflammatory response in vivo. To establish a novel link with COVID-19-related inflammatory lung injury, we followed a strategy that involved examining similar pathological situations, and we thus applied a post-influenza SA infection model that could induce significant inflammatory lung injury beyond the infection itself in mice [32], [33]. In the established mouse model, after exposure to low-dose influenza infection for 5 days, the influenza virus titer was too low to be detected; however, the immune status of the mice had been altered to “sensitive” to secondary bacterial challenge. As a result of this, severe lung inflammation accompanied by significantly elevated amounts of TNF-α and IL-6 was induced.
After influenza virus challenge for 5 days, the mice were treated with BBD (1 g/kg bw) for 2 h prior to SA infection. Survival rates, bacterial burden, pathological morphology of lung tissues, and cytokine levels of TNF-α and IL-6 in bronchoalveolar lavage fluid (BALF) were evaluated in the control (Ctrl), model (PR8 +SA), and BBD treatment groups.
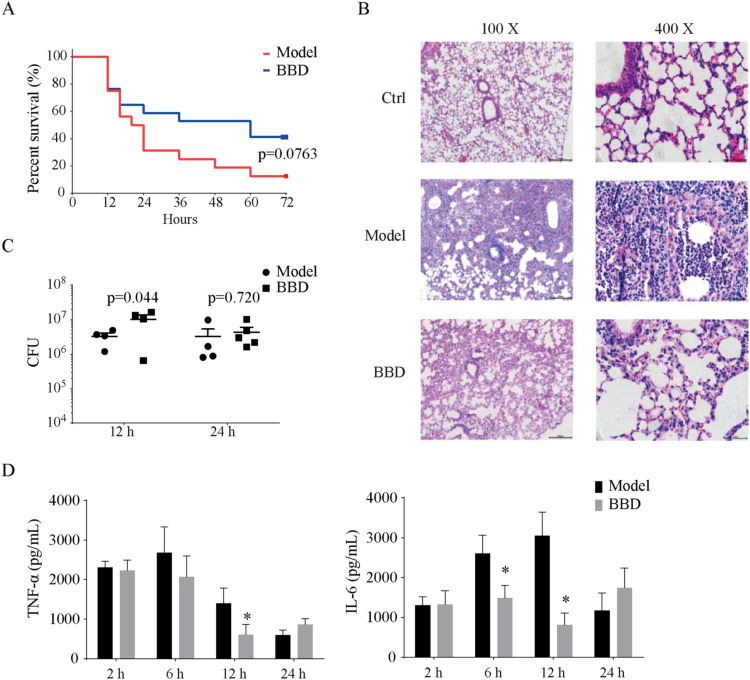
Exposure of PR8-primed mice to a high dosage (5 ×107 PFU) of SA led to the rapid death of the mice. Survival curves of the model and BBD groups were recorded. As illustrated in Fig. 6A, death occurred 12 h post SA infection in both the model and BBD groups. The mice in the model group continued to die, with only 1 out of 16 mice surviving at 72 h post SA infection. In contrast, BBD-treated mice exhibited a higher survival rate at both 48 and 72 h post SA challenge, where 7 out of 17 mice survived at 72 h post SA infection (P = 0.076).
Fig. 6.
BBD attenuates lung inflammation in mice with acute lung injury. C57BL/6 mice were infected with 200 PFU of influenza A PR/8/34 or vehicle for 5 days before being challenged with Staphylococcus aureus (SA) at the indicated CFU in 50 μL: 5 × 107 for A-B and 2.5 × 107 for C-D. (A) Survival curves; n = 16, model group; n = 17, BBD treatment group. (B) Representative histological sections of lungs at 12 h post SA challenge (n = 4–5 for each group). (C) CFU counts in LH and BAL samples (n = 4–5 for each group). (D) Concentrations of TNF-α and IL-6 in BALF (n = 3–5 for each group). * P < 0.05 vs. model group. CFU, colony-forming units; PFU, plaque-forming units; BALF, bronchoalveolar lavage fluid.
To assess the pathological features of the injured lung, we reduced the infection dosage of SA to 2.5 × 107 PFU. Microscopic histological examination of the lungs of model mice at 12 h post SA challenge revealed severe inflammatory cell infiltration and alveolar wall thickening that was indicative of acute lung injury. Treatment with BBD remarkably mitigated the lung inflammation in the model group (Fig. 6B). Nevertheless, there were no differences in bacterial load between the model and BBD groups (Fig. 6C). Moreover, the inflammatory cytokine levels of IL-6 and TNF-α in BALF samples were significantly lower in mice treated with BBD than they were in the model group. At 6 h post SA infection for IL-6 and 12 h post SA infection for both cytokines, the differences were statistically significant (P < 0.05) (Fig. 6D).
These results suggest that the anti-inflammatory effect is an important mechanism of BBD against post-influenza bacterial pneumonia. Therefore, BBD may prove to be applicable for the treatment of acute lung injury associated with severe COVID-19.
3.7. Clinical examination of the inflammatory status of seven severely ill COVID-19 patients treated with BBD
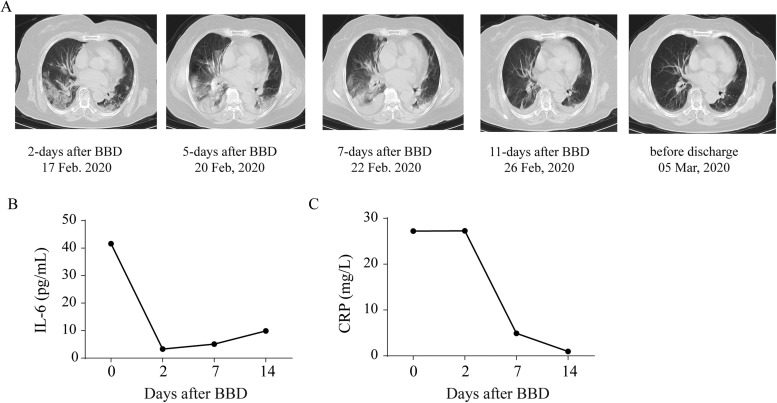
All seven patients (age range, 51–77 years; four women) met the criteria for clinical severity according to the Guideline for the Diagnosis and Treatment of COVID-19 (5th version) released by the National Health Commission of the People’s Republic of China and the National Administration of Traditional Chinese Medicine (in Chinese, available at http://www.nhc.gov.cn/xcs/zhengcwj/202002/3b09b894ac9b4204a79db5b8912d4440/files/7260301a393845fc87fcf6dd52965ecb.pdf) at the time of treatment, and all patients had received antiviral agents and methylprednisolone. The demographic characteristics of these patients are provided in Supplementary Table S7. Following BBD administration, none of the patients progressed to a critical status or required mechanical ventilation. All patients recovered and were discharged from the hospital. Changes in clinical presentations, laboratory findings, and lung CT scan results were recorded prior to treatment and after 2, 7, and 14 days of BBD treatment (Supplementary Table S8). Notably, the elimination of high amounts of IL-6 and CRP was observed in all patients. As an example, the lung CT scan images and the dynamics of IL-6 and CRP serum concentrations in patient seven (PT-7) are shown in Fig. 7.
Fig. 7.
Outcomes of CT scan image and inflammation analyses in a severely ill COVID-19 patient (PT-7) following BBD treatment. (A) Lung CT scan images for the patient at 2, 5, 7, and 11 days after BBD treatment prior to being discharged. While the bilateral pneumonia continued to develop at 2, 5, and 7 days after BBD treatment, improvement occurred on the 11th day after BBD treatment. (B-C) The dynamics of serum IL-6 and CRP levels were monitored.
4. Discussion
In our current study, we demonstrated that BBD plays a protective role against acute lung injury, thus supporting its traditional use in the treatment of infectious diseases. The NF-κB and MAPK signaling pathways are most likely related to the anti-inflammatory effects of BBD. These findings raise the possibility that BBD may prove to be helpful in the treatment of severely ill patients with COVID-19; however, this approach requires evaluation in randomized clinical trials.
The TLR1/2 and TLR3 dual activation macrophage cell model has been previously used to mimic a cytokine storm in vitro [18]. Using this model, we demonstrated that BBD possesses a powerful capacity to diminish the production of IL-6 and TNF-α. RNA-seq-based bioinformatic analyses revealed that in addition to the well-defined cytokines IL-1β, TNF-, α, and IL-6, BBD could also regulate the production of a wide spectrum of pro-inflammatory cytokines and chemokines. Furthermore, by combining transcription factor analysis and western blotting validation, we identified NF-κB and MAPK signaling pathways as targets of BBD intervention. This is not surprising, as all of these signaling pathways are involved in triggering or controlling inflammatory processes [38], [39]. Additionally, based on the results of a recent study by Tan et al. indicating that MyD88-TRIF pathways are coordinated with ERK phosphorylation for excessive pro-inflammatory cytokine production in macrophages [17], it is also likely that BBD abolishes the coordination that occurs among these signaling pathways. In the current study, we observed a dose-dependent effect for BBD. Based on this, its anti-inflammatory effect could be considered specific.
We also conducted a study incorporating the use of a post-influenza SA infection model to mimic the inflammatory status that occurs in COVID-19. As expected, after exposure to low-dose influenza infection for 5 days, the SA challenge caused severe lung inflammation due to significantly elevated amounts of TNF-α and IL-6, and this is in agreement with previous studies [32], [33]. We observed that BBD effectively protected the mice from lung injury, and the presence of abundant amounts of IL-6 and TNF-α was significantly mitigated. As the bacterial load in the mice was not altered with or without BBD treatment, the increased survival rate of mice is likely to have resulted from the inhibition of excessive immune responses in these mice.
In our case series, seven patients who were severely ill with COVID-19 were treated with BBD as a supplementary therapy to antiviral agents and methylprednisolone. As assessed by improved clinical outcomes, PaO2/FiO2 ratios, and CT scan imaging, all patients recovered from the disease and were later discharged from the hospital, and none of the patients progressed to a critically ill status or required mechanical ventilation. Importantly, the elimination of high levels of serum IL-6 and/or CRP was observed in all patients affected (PT-2, −3, −4, and −7). These results highlight the possibility that BBD may have contributed to the improvement of symptoms due to blockage of excessive inflammation. Our study does possess some limitations. First, our observations were based on a small case series that included no controls. Second, although the changes in the PaO2/FiO2 ratios and inflammatory spectrum represent encouraging findings, all the patients were treated with multiple other agents (including anti-viral medications and methylprednisolone). Based on this, large-scale randomized clinical trials are required to fully evaluate this treatment option.
It remains unknown which components of BBD are potentially responsible for the reduction in excessive host immune reactions. BBD is a TCM composed of Bovis Calculus (Niuhuang), Fel Serpentis (Shedan), Cornu Saigae Tataricae (Lingyangjiao), Margarita (Zhenzhu), Moschus (Shexiang), and Notogiseng Radix et Rhizoma (Sanqi). Li et al. applied ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry (UHPLC-Q-TOF-MS) to analyze the chemical constituents of BBD. Seventy-eight chemical compounds were identified in the categories of saponin, bile acids, and amino acids [40]. Although the bioavailability of each compound is not fully understood, the anti-inflammatory effects of Bovis Calculus (Niuhuang), Fel Serpentis (Shedan), Moschus (Shexiang), and Notogiseng Radix et Rhizoma (Sanqi) and also of saponin, bile acids, muscone, and bilirubin have been previously reported [41], [42], [43]. Using a TLR1/2 and TLR3 dual activation macrophage cell model, we identified bilirubin, muscone, and Rg3 as bioactive compounds in BBD (unpublished data). Similar to other TCM formulas, we hypothesized that the presence of multiple bioactive components in BBD could explain the simultaneous regulation of NF-κB and MAPK signaling pathways. Due to the synergistic or additive effects that occurred among the various bioactive components, BBD exhibited powerful anti-cytokine storm effects both in vivo and in vitro. Further efforts will be necessary to verify the exact molecular mode of action of BBD using a comprehensive network of pharmacological analysis-based approaches.
5. Conclusions
In conclusion, we demonstrated that BBD protects against excessive immune responses. NF-κB and MAPK signaling pathways are most likely related to the anti-inflammatory effects of BBD. These findings raise the possibility that BBD may be considered for use in treating COVID-19; however, this approach requires evaluation in randomized clinical trials.
Ethics approval and consent to participate
All animal experiments were approved by the review committee of Zhejiang University and were in compliance with institutional guidelines. The off-label clinical observations were approved by the Ethics Committee of Taizhou Hospital of Zhejiang Province, Wenzhou Medical University, Taizhou, Zhejiang, China (ChiCTR2000029769).
CRediT authorship contribution statement
KY, XF, and YW conceived and designed the study. JQ, DL, and WL performed most of the experiments. JQ and HX analyzed and interpreted the data. EC and YZ provided technical support. JQ, KY, and XF drafted the manuscript. All authors approved the final version of the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Acknowledgements
This study was supported by grants from the National S&T Major Project of China (2018ZX09201011 to XF) and the National Natural Science Foundation of China (Grant Nos. 81822047 to YW and 81970049 to KY).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2021.111586.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324:782. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020 doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pedersen S.F., Ho Y.C. SARS-CoV-2: a storm is raging. J. Clin. Investig. 2020;130:2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:84. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uccellini M.B., García-Sastre A. ISRE-reporter mouse reveals high basal and induced type I IFN responses in inflammatory monocytes. Cell Rep. 2018;25:2784–2796. doi: 10.1016/j.celrep.2018.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang D., Guo R., Lei L., Liu H., Wang Y., Wang Y., Dai T., Zhang T., Lai Y., Wang J., Liu Z., He A., O’Dwyer M., Hu J. COVID-19 infection induces readily detectable morphological and inflammation-related phenotypic changes in peripheral blood monocytes, the severity of which correlate with patient outcome. MedRxiv. 2020 doi: 10.1101/2020.03.24.20042655. 2020.03.24.20042655. [DOI] [Google Scholar]
- 11.Wen W., Su W., Tang H., Le W., Zhang X., Zheng Y., Liu X., Xie L., Li J., Ye J., Dong L., Cui X., Miao Y., Wang D., Dong J., Xiao C., Chen W., Wang H. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020 doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. Cascella, M. Rajnik, A. Cuomo, S.C. Dulebohn, R. Di Napoli, Features, Evaluation and Treatment Coronavirus (COVID-19), StatPearls, Treasure island (FL), 2020. [PubMed]
- 13.Roschewski M., Lionakis M.S., Sharman J.P., Roswarski J., Goy A., Monticelli M.A., Roshon M., Wrzesinski S.H., Desai J.V., Zarakas M.A., Collen J., Rose K.M., Hamdy A., Izumi R., Wright G.W., Chung K.K., Baselga J., Staudt L.M., Wilson W.H. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/SCIIMMUNOL.ABD0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian X., Xu F., Lung W.Y., Meyerson C., Ghaffari A.A., Cheng G., Deng J.C. Poly I:C enhances susceptibility to secondary pulmonary infections by gram-positive bacteria. PLoS One. 2012;7 doi: 10.1371/journal.pone.0041879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee B., Robinson K.M., McHugh K.J., Scheller E.V., Mandalapu S., Chen C., Di Y.P., Clay M.E., Enelow R.I., Dubin P.J., Alcorn J.F. Influenza-induced type I interferon enhances susceptibility to gram-negative and gram-positive bacterial pneumonia in mice. Am. J. Physiol. -Lung Cell. Mol. Physiol. 2015;309:L158–L167. doi: 10.1152/ajplung.00338.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shepardson K.M., Larson K., Morton R.V., Prigge J.R., Schmidt E.E., Huber V.C., Rynda-Apple A. Differential type I interferon signaling is a master regulator of susceptibility to postinfluenza bacterial superinfection. MBio. 2016;7 doi: 10.1128/mBio.00506-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ting Tan R.S., Lin B., Liu Q., Tucker-Kellogg L., Ho B., Leung B.P., Ding J.L. The synergy in cytokine production through MyD88-TRIF pathways is co-ordinated with ERK phosphorylation in macrophages. Immunol. Cell Biol. 2013;91:377–387. doi: 10.1038/icb.2013.13. [DOI] [PubMed] [Google Scholar]
- 18.Lin B., Dutta B., Fraser I.D.C. Systematic investigation of multi-TLR sensing identifies regulators of sustained gene activation in macrophages. Cell Syst. 2017;5:25–37. doi: 10.1016/j.cels.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020 doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 20.Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J., Kui Z., Shuxiang H., Jun D., Xiaobo L., Xiaotao H., Lin W., Nanshan Z., Zifeng Y. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q., Pang P., Zheng K., Sun L., Wang J., Chen X. Xin-Jia-Xiang-Ru-Yin alleviated H1N1-induced acute lung injury and inhibited the IFN-gamma-related regulatory pathway in summer flu. Biomed. Pharmacother. 2018;108:201–207. doi: 10.1016/j.biopha.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 22.Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., Zheng M., Zhang X., Xia D., Ke Y., Lu L., Wang D. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:802–816. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y., Tian Z.Q., Liang L., Yang X., Sheng D.D., Zeng J.X., Li X.Y., Shi R.Y., Han Z.P., Wei L.X. Babao Dan attenuates acute ethanol-induced liver injury via Nrf2 activation and autophagy. Cell Biosci. 2019;9:80. doi: 10.1186/s13578-019-0343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheng D., Zhao S., Gao L., Zheng H., Liu W., Hou J., Jin Y., Ye F., Zhao Q., Li R., Zhao N., Zhang L., Han Z., Wei L. BabaoDan attenuates high-fat diet-induced non-alcoholic fatty liver disease via activation of AMPK signaling. Cell Biosci. 2019;9:77. doi: 10.1186/s13578-019-0339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu L., Wu C., Lu B.J., Xie D., Wang Z., Azami L., An Y.T., Wang H.J., Ye G., Sun M.Y. Babaodan cures hepatic encephalopathy by decreasing ammonia levels and alleviating inflammation. J. Ethnopharmacol. 2019 doi: 10.1016/j.jep.2019.112301. [DOI] [PubMed] [Google Scholar]
- 26.Gong W., Liu L., Li M., Wang L., Zhang M., Luo Z., Sridhar S., Woo P.C.Y., Wang L. Evaluation of antiviral efficacy of Chinese traditional medicine Babao Dan in rabbits infected with hepatitis E virus. J. Gen. Virol. 2018;99:1036–1043. doi: 10.1099/jgv.0.001089. [DOI] [PubMed] [Google Scholar]
- 27.Liang L., Yang X., Yu Y., Li X., Wu Y., Shi R., Jiang J., Gao L., Ye F., Zhao Q., Li R., Wei L., Han Z. Babao Dan attenuates hepatic fibrosis by inhibiting hepatic stellate cells activation and proliferation via TLR4 signaling pathway. Oncotarget. 2016;7:82554–82566. doi: 10.18632/oncotarget.12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q., Liu Z., Du K., Liang M., Zhu X., Yu Z., Chen R., Qin L., Li Y., Zheng Y. Babaodan inhibits cell growth by inducing autophagy through the PI3K/AKT/mTOR pathway and enhances antitumor effects of cisplatin in NSCLC cells. Am. J. Transl. Res. 2019;11:5272–5283. 〈https://www.ncbi.nlm.nih.gov/pubmed/31497240〉 [PMC free article] [PubMed] [Google Scholar]
- 29.Du Y., Xin L., Shi Y., Zhang T.H., Wu N.C., Dai L., Gong D., Brar G., Shu S., Luo J., Reiley W., Tseng Y.W., Bai H., Wu T.T., Wang J., Shu Y., Sun R. Genome-wide identification of interferon-sensitive mutations enables influenza vaccine design. Science. 2018;359:290–296. doi: 10.1126/science.aan8806. (80-.) [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Hong Z., Qian J., Wang Y., Wang S. Protective effect of Jie-Geng-Tang against Staphylococcus aureus induced acute lung injury in mice and discovery of its effective constituents. J. Ethnopharmacol. 2019;243 doi: 10.1016/j.jep.2019.112076. [DOI] [PubMed] [Google Scholar]
- 31.Qian J., Hu Y., Zhao L., Xia J., Li C., Shi L., Xu F. Protective role of adipose-derived stem cells in staphylococcus aureus-induced lung injury is mediated by RegIIIgamma secretion. Stem Cells. 2016;34:1947–1956. doi: 10.1002/stem.2337. [DOI] [PubMed] [Google Scholar]
- 32.Smith A.M., Adler F.R., Ribeiro R.M., Gutenkunst R.N., McAuley J.L., McCullers J.A., Perelson A.S. Kinetics of coinfection with influenza A virus and Streptococcus pneumoniae. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Li F., Sun R., Gao X., Wei H., Li L.J., Tian Z. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat. Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gotea V., Ovcharenko I. DiRE: identifying distant regulatory elements of co-expressed genes. Nucleic Acids Res. 2008;36:W133–W139. doi: 10.1093/nar/gkn300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmolke M., Viemann D., Roth J., Ludwig S. Essential impact of NF-kappaB signaling on the H5N1 influenza A virus-induced transcriptome. J. Immunol. 2009;183:5180–5189. doi: 10.4049/jimmunol.0804198. [DOI] [PubMed] [Google Scholar]
- 39.Droebner K., Reiling S.J., Planz O. Role of hypercytokinemia in NF-kappaB p50-deficient mice after H5N1 influenza A virus infection. J. Virol. 2008;82:11461–11466. doi: 10.1128/JVI.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Yu J., Zhao Y.H., An Y.T., Xue D., Chai Y.F., Zhang G.Q., Zhang H. UHPLC-Q-TOF-MS in analyzing chemical constituents of traditional Chinese herbal preparation Babao Dan. Acad. J. Second Mil. Univ. 2016;37:1548–1554. doi: 10.16781/j.0258-879x.2016.12.1548. [DOI] [Google Scholar]
- 41.Guo C., Chi Z., Jiang D., Xu T., Yu W., Wang Z., Chen S., Zhang L., Liu Q., Guo X., Zhang X., Li W., Lu L., Wu Y., Song B.L., Wang D. Cholesterol homeostatic regulator SCAP-SREBP2 integrates NLRP3 inflammasome activation and cholesterol biosynthetic signaling in macrophages. Immunity. 2018;49:842–856. doi: 10.1016/j.immuni.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 42.Guo C., Xie S., Chi Z., Zhang J., Liu Y., Zhang L., Zheng M., Zhang X., Xia D., Ke Y., Lu L., Wang D. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:802–816. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 43.Li Y., Huang B., Ye T., Wang Y., Xia D., Qian J. Physiological concentrations of bilirubin control inflammatory response by inhibiting NF-κB and inflammasome activation. Int. Immunopharmacol. 2020;84 doi: 10.1016/j.intimp.2020.106520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material