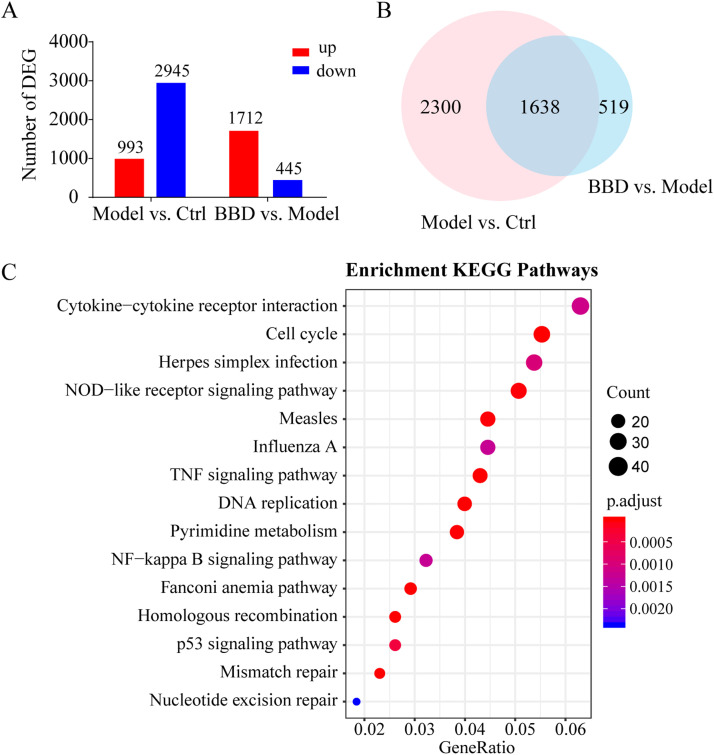
Fig. 3.
Gene expression profiling by RNA-seq. Raw 264.7 cells grown on 6-well plates were treated with a combination of 10 μg/mL Poly(I:C) and 100 ng/mL Pam3CSK4 (model) for 8 h with or without pre-treatment with 1 mg/mL of Babaodan (BBD) for 24 h. RNA samples were collected for RNA-seq analysis. Two samples were analyzed per group. Differentially expressed genes (DEG) are identified by absolute log2 (FC) > 2 and P < 0.05 values. (A) Number of up- and downregulated genes in the model and treatment groups. (B) Venn diagram summary of DEGs between the control vs. model and model vs. BBD groups. (C) Top 15 KEGG pathway enrichment scores of co-regulated DEGs in the model vs. control and BBD vs. model groups.