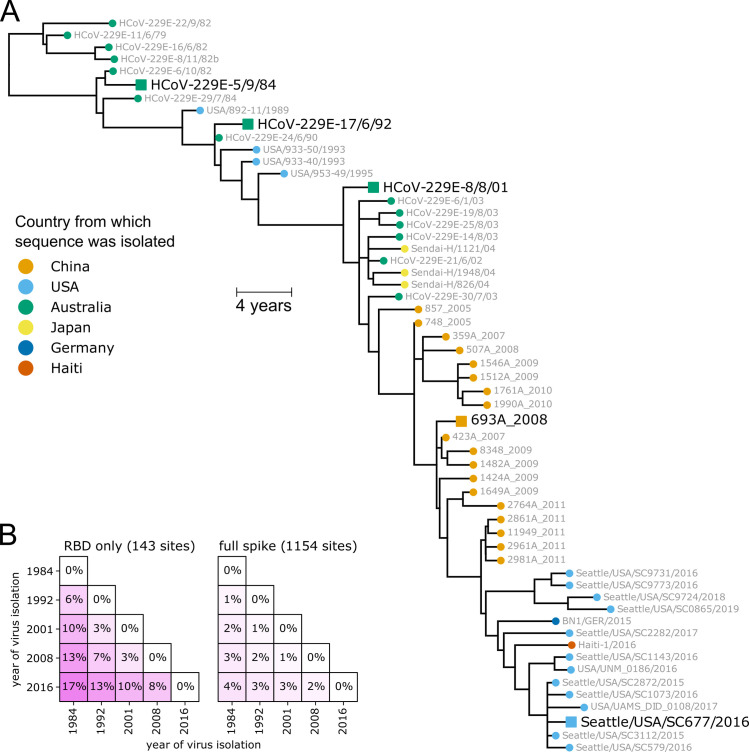
Fig 1. Spikes used in this study.
(A) Phylogenetic tree of 229E spikes, with tips colored by the country from which the virus was isolated. The spikes used in the experiments are indicated with black text and square shapes. The tree is a maximum-likelihood inference with IQ-TREE [82] with a codon-substitution model and re-scaled with TreeTime [84] to position tips by to date of isolation. S1 Fig shows a tree with branch lengths proportional to divergence rather than time, and validates clock-like evolution. S2 Fig shows recombination does not substantially affect the phylogenetic placements of the spikes used in the experiments. (B) Protein sequence divergence of the spikes used in the experiments, computed over just the receptor-binding domain (RBD) or the full sequence. Divergence is the Levenshtein distance between the amino-acid sequences divided by the number of sites.