Figure 1.
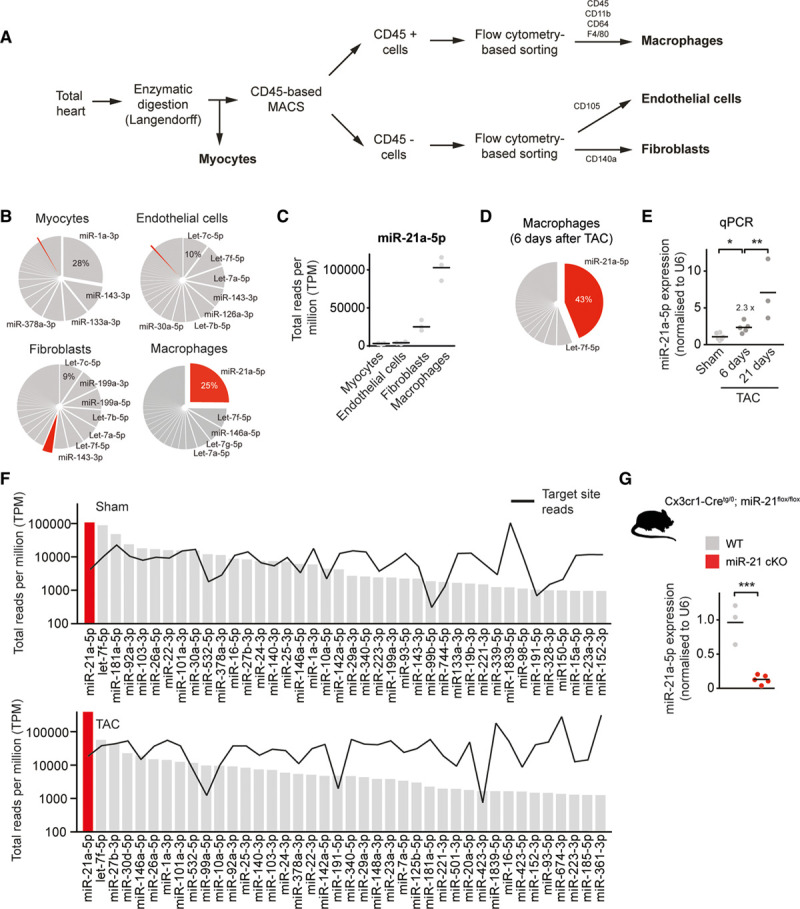
miR-21 is the most abundant and enriched microRNA in cardiac macrophages. A, Scheme for cell isolation. B, MiRNA expression profiles in different myocardial cell types. Pie graph representation of miRNAs in cardiac macrophages and other myocardial cell types (myocytes, fibroblasts, and endothelial cells) isolated from hearts of WT mice. miR-21a-5p is indicated in red. n=3. C, Expression of miR-21a-5p in different myocardial cell types. D, Pie graph representation of miRNAs in cardiac macrophages isolated from hearts of WT mice 6 days after pressure overload (TAC). n=5. E, Relative expression of miR-21a-5p, as determined by SYBR green-based quantitative real-time polymerase chain reaction (qPCR) in cardiac macrophages isolated from hearts 6 or 21 days after pressure overload. Sham n=6; 6 days after TAC n=5; and 21 days after TAC n=3. F, Measured reads of miRNA (bars) and corresponding 3′-untranslated region target pools (black line) in total reads per million (TPM) for the top-expressed miRNAs in cardiac macrophages, isolated from WT mice after sham or TAC surgery. MiR-21 is denoted in red. The y axis is log scale. n=3 per group. G, Macrophage-specific miR-21–deficient mice (miR-21 cKO) were generated by crossing miR-21–floxed mice with a mouse line that expresses Cre recombinase under control of the Cx3cr1 promotor. Diagram shows the expression of miR-21a-5p as determined in macrophages isolated from WT and miR-21 cKO mice. WT n=3; miR-21 cKO n=5. Data denote mean and individual values and were analyzed using 1-way ANOVA (E) or the Student t test (G). *P<0.05, **P<0.01, and ***P<0.001. MACS indicates magnetic-activated cell sorting; miR-21, microRNA-21; miR-21 cKO, macrophage-specific miR-21–deficient mice; miRNA, microRNA; TAC, transverse aortic constriction; and WT, wild type.
