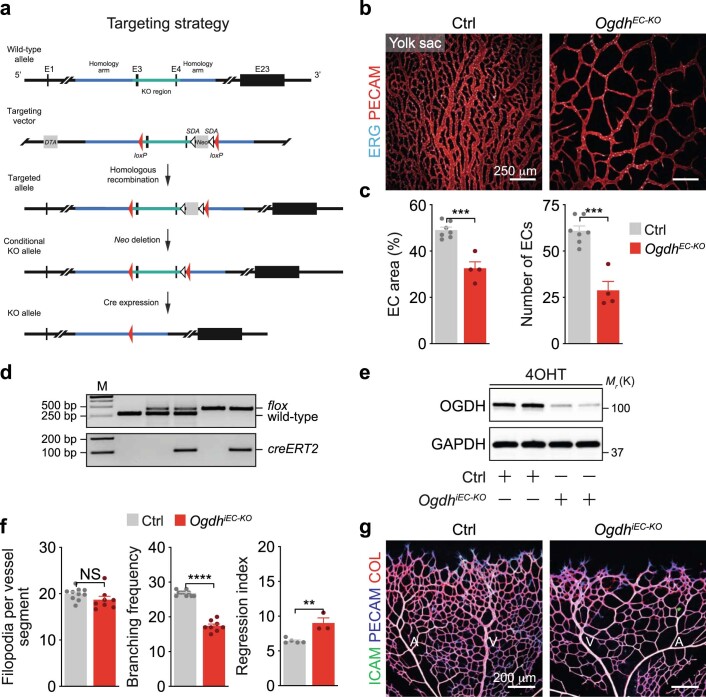
Extended Data Fig. 7. Loss of endothelial Ogdh impairs vascular growth.
a, Schematic illustration of the strategy to generate a conditional Ogdh knockout allele, in which exons 3 and 4 are flanked by loxP sites (red triangles). The Ogdh genomic locus, the targeting vector, the targeted allele and the SDA- and cre-recombined loci are shown. SDA-Neo-SDA, neomycin resistance cassette flanked by SDA sites. b, Immunofluorescence staining for ERG (cyan) and PECAM1 (PECAM, red) of control (Ctrl, Ogdhfl/fl) and endothelial-restricted Ogdh knockout (OgdhEC-KO, Tie2-cre;Ogdhfl/fl) yolk sacs at E11.5, showing reduced vascular density after Ogdh loss. c, Quantifications of vascular parameters in E11.5 Ctrl and OgdhEC-KO yolk sacs, as indicated, (EC area: n=7, 4 samples for Ctrl and OgdhEC-KO; Number of ECs: n=7, 4 samples for Ctrl and OgdhEC-KO). d, PCR analysis of genomic DNA from control (Ogdh+/+, lane 2; Ogdhfl/+, lane 3; Ogdhfl/fl, lane5) and conditional Ogdh mutant mice (Cdh5-creERT2; Ogdhfl/+, lane4; Cdh5-creERT2;Ogdhfl/fl, lane 6). Lane 1, DNA marker (M). e, Immunoblot analysis of OGDH protein levels in ECs isolated from the liver of 4-OHT-injected Ctrl (Ogdhfl/fl) and OgdhiEC-KO (Cdh5-creERT2;Ogdhfl/fl) mouse mutants. GAPDH served as loading control. f, Quantifications of vascular parameters in P6 Ctrl and OgdhiEC-KO mouse mutants, as indicated (Filopodia per vessel segment: n=10, 8 samples for Ctrl and OgdhiEC-KO; Branching frequency: n=10, 8 samples for Ctrl and OgdhiEC-KO; Regression index: n=5, 3 samples for Ctrl and OgdhiEC-KO). g, Immunostaining showing ICAM2- (ICAM, green), PECAM- (blue), and collagen IV- (COL, red) labelling of P6 retinas from 4-OHT-injected Ctrl and OgdhiEC-KO mice. Western blot data in e were from the respective experiment, processed in parallel, and are representative of at least three independent experiments. c and f, Data represent mean ± s.e.m.; a two-tailed unpaired t-test was used; ** P< 0.01; ***P<0.001; ****P<0.0001; NS, not significant. The numerical data, unprocessed western blots and P values are provided as source data.