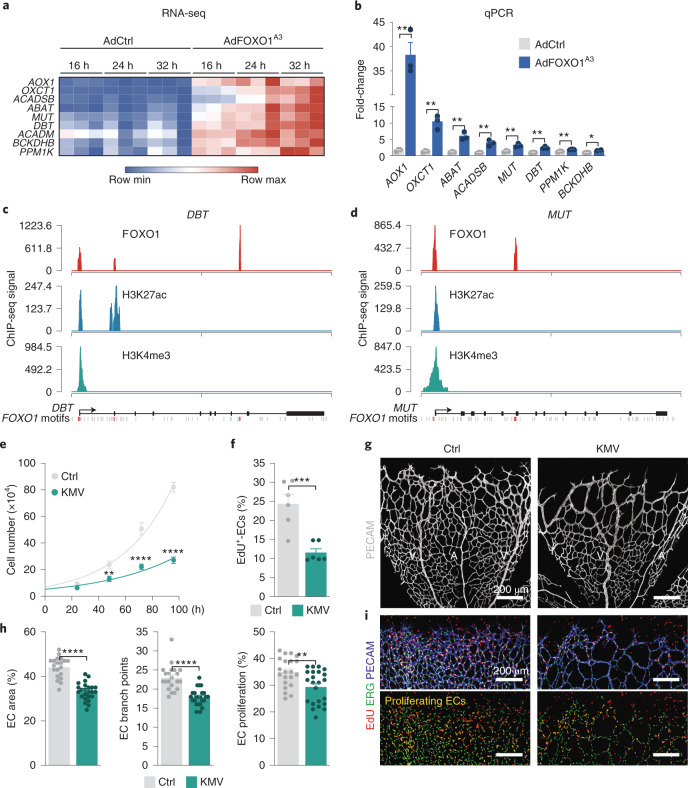
Fig. 7. The FOXO1-regulated BCAA catabolite KMV limits endothelial proliferation.
a, Heatmap of genes involved in BCAA metabolism that are induced in FOXO1A3-expressing (AdFOXO1A3) HUVECs compared to control (AdCtrl). Transcript levels were assessed at 16, 24 and 32 h post-transduction and analysed by RNA-seq. Genes with a fold-change ≥2 and a P value of <0.05 are shown (n = 3 independent samples). b, RT–qPCR analysis validating the increased expression of BCAA metabolism genes in AdFOXO1A3-transduced HUVECs. Values are normalized to β-actin levels and represented as fold-change relative to control (n = 3 independent samples). c,d, FOXO1, H3K27ac and H3K4me3 ChIP-seq signals at the genomic loci of the DBT (c) and MUT (d) gene. FOXO consensus motifs bound by FOXO1 are indicated in orange. Unbound FOXO motifs are shown in grey. ChIP-seq signals are represented as reads per kilobase million. e, Cell-proliferation curves of HUVECs treated with vehicle (PBS, Ctrl) or KMV for the indicated times (n = 12 independent samples). f, EdU incorporation is reduced in HUVECs treated with KMV for 48 h (n = 6 independent samples). g, Confocal images for PECAM (grey) labelling in P7 mouse retinas showing decreased vascular density after a single intraocular injection of KMV at P5. Mice injected with PBS were used as a control. h, Quantifications of vascular parameters in the retina of control and KMV-injected mice, as indicated (EC area: n = 20 (control) and 24 (KMV) samples; EC branch points: n = 21 (control) and 24 (KMV) samples; EC proliferation: n = 21 (control) and 23 (KMV) samples). i, EdU (red), ERG (green) and PECAM (blue) labelling of control and KMV-injected retinas at P7, revealing decreased endothelial proliferation following KMV treatment. For b, e, f and h, the data represent the mean ± s.e.m.; two-tailed unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The numerical data and P values are provided as source data.