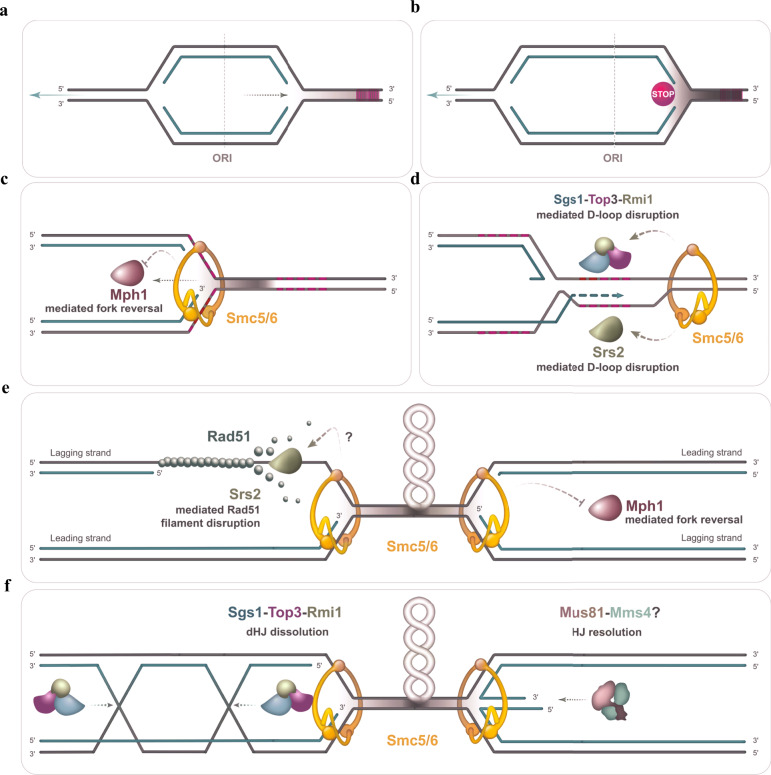
Fig. 8. Schematic representation of Smc5/6 roles in coordinating STR and other DNA crossed strand processing enzymes upon pausing of replication forks at natural pause sites.
The upper panel shows replication forks progressing normally (a) and one fork pausing as it begins traversing elements at a NPS shown in dashed red lines (b). Upon fork stalling, Smc5/6 is shown to inhibit the fork regression activity of Mph1 (c) and to promote Srs2 and Sgs1–Top3–Rmi1-mediated disruption of displacement loops (D-loops) (d). In panel d, the D-loops are shown to form by toxic strand invasion, presumably generated upon fork reversal, into repetitive regions (shown as dashed red lines) present in the non-replicated DNA ahead of the stalled fork. As replication forks converge, Smc5/6 may act to manage associated topological stress and prevent formation of recombination intermediates and reversed forks by regulating Srs2 and Mph1, respectively (e). At converging forks, in the context of matured recombination structures (f), Smc5/6 is shown to facilitate STR complex in processing double Holliday Junctions (dHJs) and Mus81–Mms4 in resolving reversed forks, resembling single Holliday Junctions.