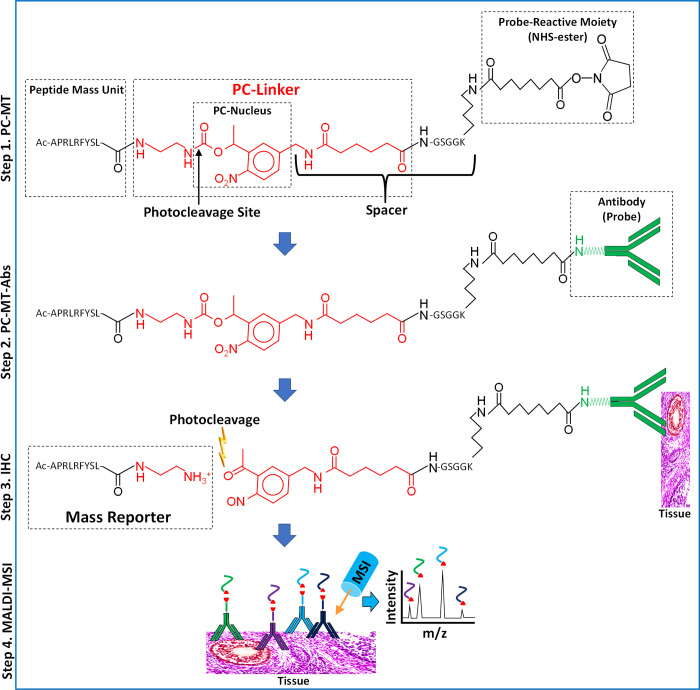
Figure 1.
Structure and use of PC-MTs and PC-MT antibodies (PC-MT-antibodies). (Step 1, PC-MT) The structure of a PC-MT is shown. To produce the PC-MTs, an amine-terminal Fmoc-protected version of the photocleavable linker (PC-Linker; red) is incorporated during conventional Fmoc-based solid-phase peptide synthesis (SPPS) along with the other amino acids. The example PC-MT shown is oriented with the N-terminal on the left and the C-terminal on the right. “APRLRFYSL” is an example amino acid sequence of the peptide mass unit (see Supplementary Table S1 for all mass units used in this work). The PC-MTs contain a spacer that connects the 1-(2-nitrophenyl)-ethyl-based photocleavable nucleus (PC-Nucleus) to the probe-reactive moiety. This spacer is comprised of a portion of the PC-Linker plus the GSGGK amino acid sequence. The probe-reactive moiety (an NHS-ester leaving group) is generated during SPPS on the ε-amine of the lysine (K) included in the spacer. N-Terminal acetylation (“Ac”) of the α-amine is used to prevent the self-reaction and polymerization of the PC-MT. (Step 2, PC-MT-antibodies) The NHS-ester probe-reactive moiety of the PC-MTs is then reacted in one step with native primary amines in the antibody probes to form the PC-MT antibodies (PC-MT-antibodies). (Step 3, IHC) Immunohistochemistry (IHC) is performed by binding the PC-MT-antibodies to targets in thin tissue sections mounted on conductive microscope slides, followed by photocleavage under dry conditions to liberate the mass reporters. Note that a small residual portion of the PC-Linker (red) remains as part of the photocleaved mass reporters. (Step 4, MALDI-MSI) Finally, MALDI mass spectrometric imaging (MSI) is performed following the application of a matrix compound (matrix not shown). The inverted Y-shaped structures are antibodies, and the curved colored lines correspond to the mass units from different PC-MTs. The red shapes correspond to the photocleaved PC-Linker.