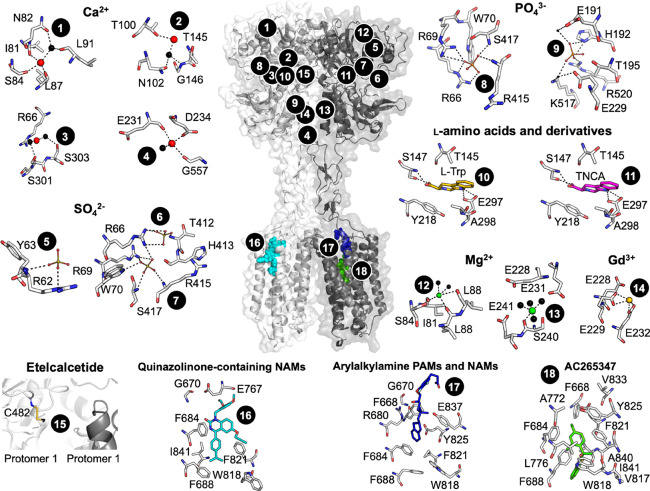
Figure 2.
CaSR model and predicted ligand binding sites. The published crystal structure of the CaSR ECD (PDB 5K5S) was superimposed onto a published model of the CaSR 7TM, ECLs, and ICLs71 based on homology with the mGlu5 crystal structure (PDB 6N51). Numbers correspond to ligand binding sites predicted as follows: Ca2+ (sites 1–4) by anomalous scattering analysis and SO42– (5–7), PO43– (8–9), and l-Trp (10) by electron density distribution analysis of the crystallized ECD (PDBs 5K5S and 5K5T);31 TNCA (11), Mg2+ (12–13), and Gd3+ (13) by electron density distribution analysis of the crystallized VFT (PDBs 5FBK and 5FBH);33 etelcalcetide (15) from mutagenesis and mass spectrometry, where the yellow stick represents a putative disulfide bond as a rough depiction of where etelcalcetide is predicted to bind;66 quinazolinone-containing NAMs (16), arylalkylamine PAMs and NAMs (17), and AC265347 (18) from mutagenesis combined with homology modeling and computational docking.41,71