Figure 4. Imatinib, mycophenolic acid, and quinacrine dihydrochloride each block the entry of SARS-CoV-2 virus in both hPSC-derived LOs and COs.
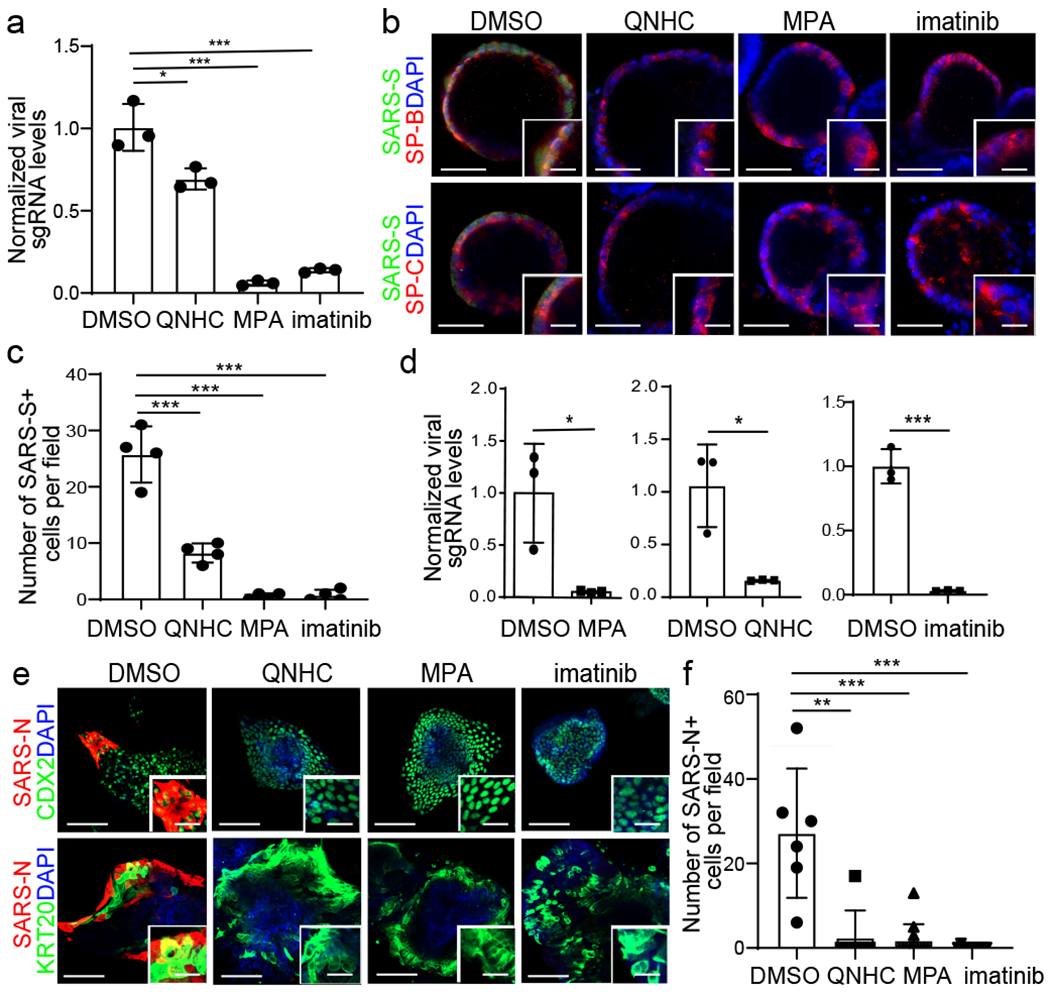
a-c, qRT-PCR analysis of total RNA extracted from infected hPSC-LOs for viral N sgRNA (a, n=3 biological independent experiments, *P=0.0256, ***P=0.000333, ***P=0.000461), immunofluorescent staining (b) and quantification (c, n=4 biological replicates for each group, ***P=6.36E-05, ***P=5.63E-05, ***P=0.000566) of SARS-CoV-2 Spike protein (SARS-S) and SP-B/SP-C in imatinib, MPA, or QNHC treated hPSC-LOs at 24 hpi (MOI=0.5). Scale bar= 50 μm. Microscale bar= 15 μm. d-f, qRT-PCR analysis of total RNA extracted from infected hPSC-COs for viral N sgRNA (d, n=3 biological independent experiments, ***P=0.0260, ***P=0.0166, ***P=0.000235), immunofluorescent staining (e) and quantification (f, n=6 biological independent experiments, **P=0.00242, ***P=4.34E-05, ***P=5.26E-05) of SARS-CoV-2 nucleocapsid protein (SARS-N) of imatinib, MPA, or QNHC-treated hPSC-COs at 24 hpi (MOI=0.5). Scale bar= 50 μm. Microscale bars=15 μm. *P < 0.05, **P < 0.01, and ***P < 0.001. Data were analyzed by an unpaired two-tailed Student’s t-test and shown as mean ± STDEV. Data are representative of at least three independent experiments.
