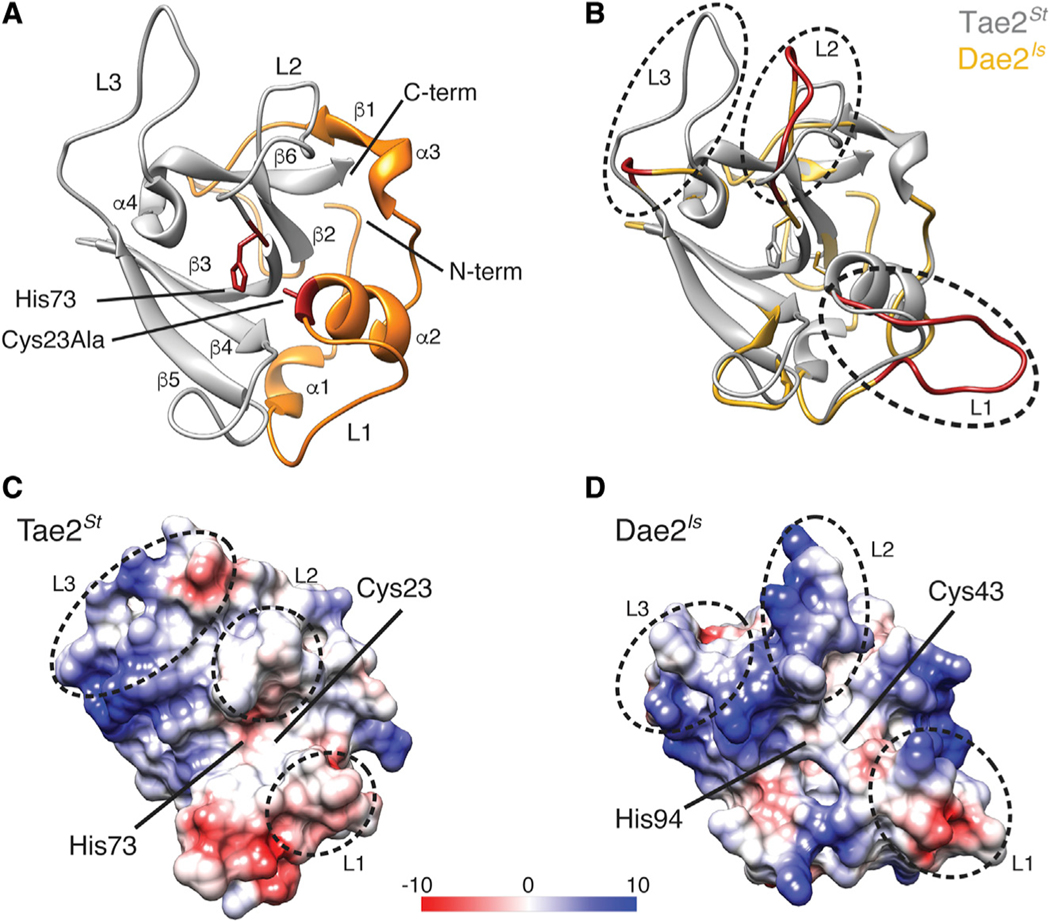
Figure 1. Tae2 and Dae2 Enzymes Have Divergent Structural Features.
(A) Ribbon diagram Tae2 from Salmonella enterica Typhi (Tae2St). Canonical papain-like secondary structures are labeled (α1–4, β1–6, Loop1–3 [L1–3]), and subdomains are colored (gray, orange). Catalytic residues are denoted (labeled His 73 and Cy-s23Ala, red).
(B) Structural alignment of Tae2St (gray) and a Tae2-based homology model of Dae2Is (golden). Loops 1, 2, and 3 are colored red in Dae2Is.
(C and D) Electrostatic charges in surfaces proximal to the catalytic residues for Tae2St and Dae2Is (scale is in kcal mol−1e−1). The loops are boxed in sequence alignment in Figure S1B.