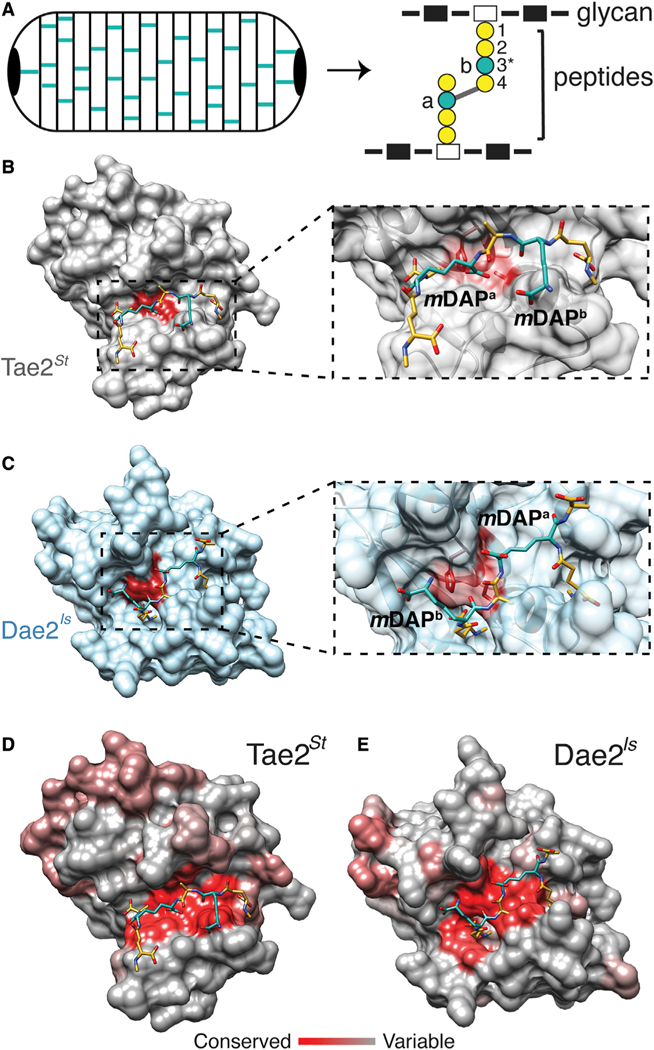
Figure 2. Modeling of Tae2/Dae2 Interactions with PG Substrate.
(A) Cartoon schematic of bacterial peptidoglycan (PG) sacculus (left) and PG chemical structure (right) with glycans (rectangles), peptides (circles), and cross-link position (gray line) shown. Residue positions in PG peptide stem are labeled (1–4) with key variable third position (3*) colored in teal.
(B) Surface representation of Tae2St ligand docking using a stem peptide substrate. Both mDAP residues in the stem peptide are colored in teal.
(C) Close-up of the Tae2St active site and docked substrate. The scissile amide bond is between mDAPa and the adjacent D-ala (position 4), proximal to the active site Cys23 residue.
(D) Surface representation of Dae2Is ligand docking, using the same substrate as for Tae2St in (C).
(E) Close-up of the Dae2Is active site and docked substrate. The scissile bond is similarly situated between mDAPa and the adjacent D-ala, proximal to the active site Cys43 residue. The substrate directionality being mDAPb – D-ala – mDAPa is the inverse of that for Tae2St, mDAPa – D-ala – mDAPb.
(F) Consurf server analyses of Tae2St and Dae2Is with highly conserved residues shown in red and least conserved in gray. The docked PG substrate is shown.
See also Figures S3 and S4.