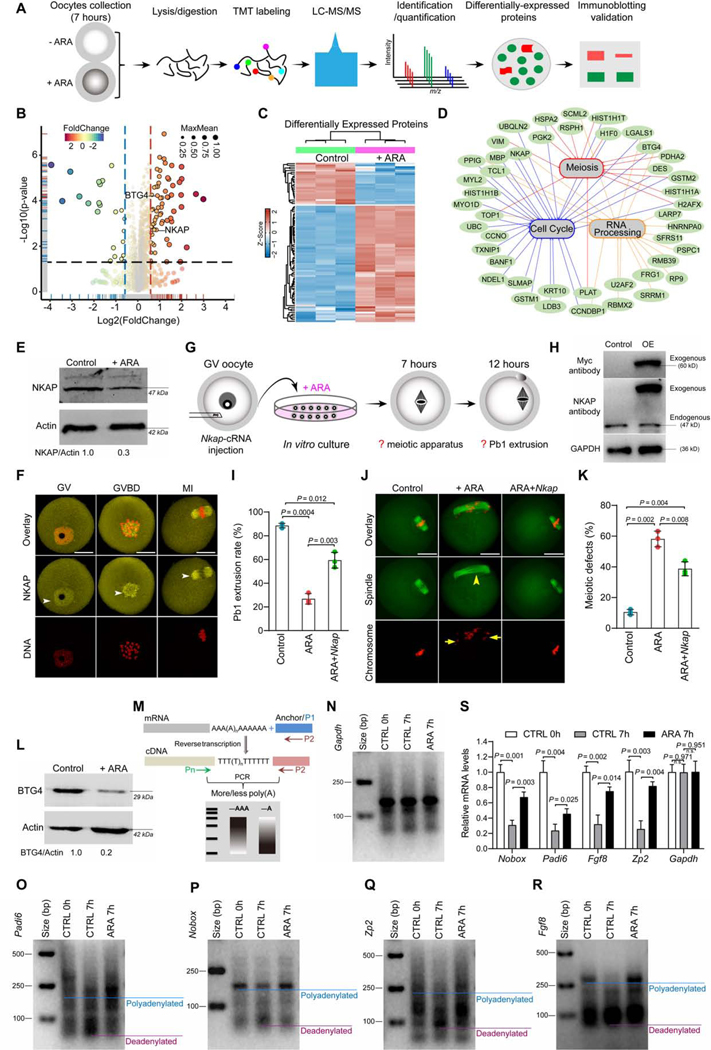
Figure 3. Identification of factors mediating the effects of ARA on oocyte maturation.
(A) Overview of the proteomic workflow for identifying differential protein accumulation. (B) Volcano plot showing the relative abundance of proteins. (C) Heat map of the differentially accumulated proteins between control and ARA-treated oocytes. (D) Gene ontology network enrichment analysis of the proteins with differential accumulation. (E) Western blot analysis of NKAP expression validated the proteomic results (200 oocytes per lane). (F) Cellular distribution of NKAP in oocytes at GV, GVBD and MI stages. Arrowheads point to NKAP signals. Scale bars, 25 μm. (G) Schematic presentation of the experimental protocol to check whether NKAP overexpression could suppress the defective phenotypes of ARA-treated oocytes. (H) Immunoblotting showing the overexpression (OE) of exogenous NKAP protein in oocytes. (I) Incidence of Pb1 extrusion in indicated oocytes. (J) Representative examples of meiotic spindle and chromosomes in indicated oocytes. Spindle disorganization and chromosome misalignment are indicated by arrowheads and arrows, respectively. Scale bars, 25 μm. (K) Quantitative analysis of meiotic defects in indicated oocytes. At least 100 oocytes for each group were analyzed, and the experiments were conducted three times. (L) Western blot analysis of BTG4 expression validated the proteomic results (200 oocytes per lane). (M) Diagram showing the strategy of the mRNA poly(A) tail (PAT) assay. P1, anchor primer; P2, P1-antisense primer; Pn, gene-specific primer. (N-R) PAT assay showing changes in the poly(A)-tail length for the indicated transcripts in control and ARA-treated oocytes. (S) Relative abundance of the indicated transcripts in control and ARA-treated oocytes, determined by real-time RT-PCR. Error bars, SD. Student’s t test was used for statistical analysis in all panels. n.s., not significant. See also Figure S8 and Table S3.