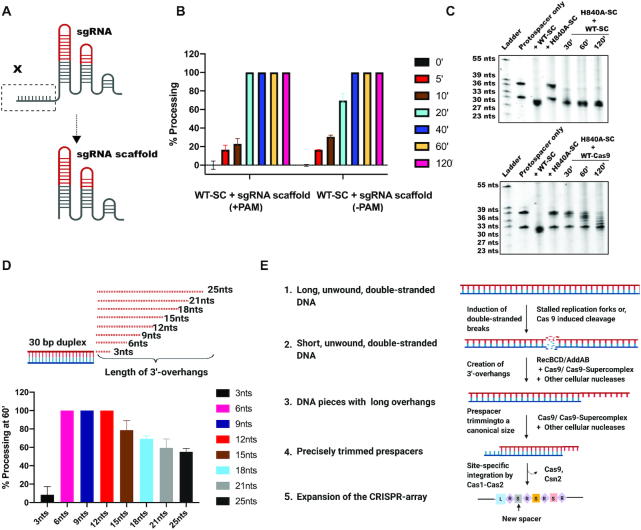
Figure 5.
Prespacer processing by supercomplex is independent of crRNA spacer sequence. (A) Schematic showing the design of sgRNA scaffold. (B) WT-supercomplex, in complex with sgRNA-scaffold, trims PAM-containing prespacers with relatively faster kinetics. Bar graphs represent the percentage processing with time, for prespacers either containing or lacking a PAM. Error bars represent ± SD; n = 3. (C) Rescue of in vitro prespacer processing by supplementing H840A-supercomplex with either WT-supercomplex or WT-Cas9. (D) Design of substrates with different lengths of 3′-overhangs for assessing substrate preference by supercomplex (top). Quantification of processing by the WT-supercomplex for prespacers containing different 3′-overhang lengths are represented as bar graphs (bottom); n = 3 and error bars represent ± SD. (E) Model of prespacer processing by the Cas9-integration supercomplex. Self-derived spacers are most frequently derived from genomic loci with a high frequency of generating double-stranded breaks. In S. pyogenes, such double-stranded breaks are repaired by AddAB, which creates DNA pieces with long 3′-overhangs. These products can be captured by the Cas9-supercomplex along with additional cellular nucleases and processed to a canonical size for integration by the Cas1-Cas2 integrase for site-specific integration.