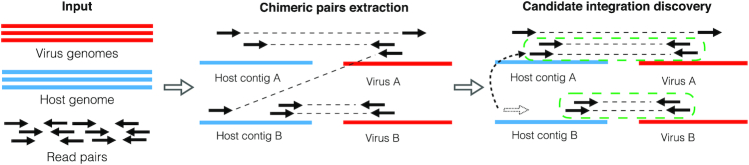
Figure 2.
SurVirus pipeline. The input to the algorithm is a host genome, a database of virus sequences and a set of read pairs. The algorithm then proceeds in two steps. Step 1: Chimeric pairs extraction. It extracts the subset of read pairs that are useful to the prediction of virus integrations, and maps them onto the human and viruses genomes using a standard aligner. Step 2: Candidate integration discovery. It corrects the alignment of the reads that were incorrectly aligned (for example, in the figure, the read represented as dotted arrow is realigned to the correct location), and clusters the pairs to predict virus integrations. Each cluster represents the breakpoint of an integration. (For example, in the figure, we discovered two clusters which are enclosed by green dotted lines.)