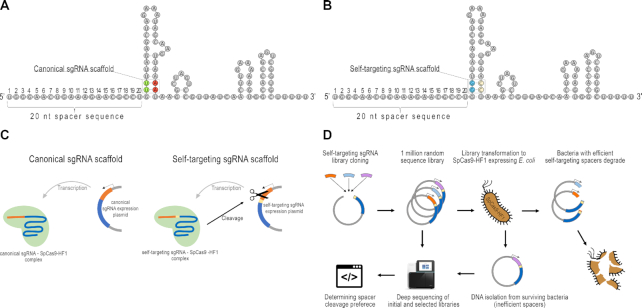
Figure 1.
Principle of the Self-targeting sgRNA Library Screen (SLS) method. (A, B) Representation of sgRNAs with canonical and self-targeting scaffolds. Position 22–23 (TT dinucleotides, (green) (A) in the coding sequence of the sgRNA are altered to GG (blue) to generate a PAM motif, resulting in the construction of a self-targeting sgRNA (B). To maintain the stem structure, the complement strand is also modified. (C) Self-targeting sgRNAs, in contrast to canonical sgRNAs, target their own coding DNA sequence. The spacer sequence is shown in orange, the PAM in yellow, the sgRNA scaffold in blue, the plasmid backbone in gray. (D) Flowchart of the SLS method developed to identify SpCas9-HF1’s spacer preference. The plasmids harboring effectively-cleaved target sequences derived from the one million sequence library are depleted under antibiotic selection. Sequencing both the initial and the nuclease-processed libraries, cleaved and uncleaved spacer sequences can be identified.