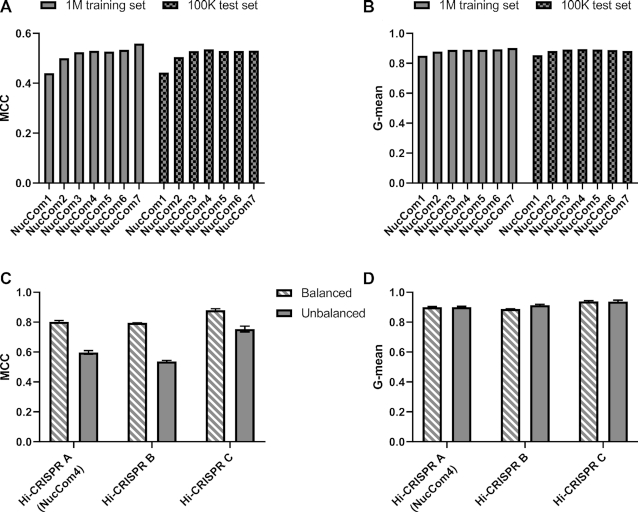
Figure 2.
Prediction performance of Hi-CRISPR predictors on target-library data. (A, B) Binary classification results on the 1M training (plain bars) and on the 100K test libraries (chequered bars). NucCom 1–7 are position-dependent short (one to seven base long) motif-based predictors developed using the 1M library data. The quality of the classifications is assessed by the Matthews correlation coefficient (MCC) (A) and G-mean (i.e. the geometric average of sensitivity and specificity) (B). Increasing the lengths of the motifs up to four or seven nucleotides improves the G-mean and the MCC values of the predictions, respectively, on the 100K test libraries. (C, D) SpCas9-HF1 predictions developed in this study are compared on three, either balanced (50% efficiently cleaved, 50% weakly cleaved spacers – striped bars) or unbalanced (93.4% efficiently cleaved, 6.6% weakly cleaved – plain bars) target pools randomly selected from the 100K test dataset. Hi-CRISPR A (NucCom4 shown on A and B), Hi-CRISPR B (Deep-learning scheme based on (49)) and Hi-CRISPR C (Deep-learning scheme based on (29)). MCC (C), but not the G-mean (D) values are sensitive to whether balanced (striped bars) or unbalanced (plain bars) datasets are used. Columns represent means ± SD of the predictions on the three datasets.