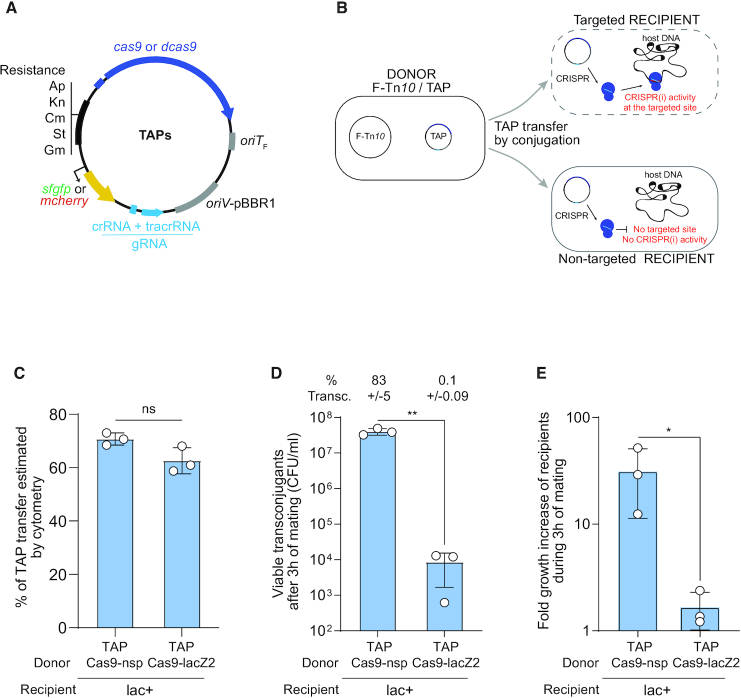
Figure 1.
Transfer of TAP by F plasmid machinery mediates killing of a targeted E. coli strain. (A) TAPs modules consist of CRISPR system composed of wild type (cas9) or catalytically dead cas9 (dcas9) genes expressed from the weak constitutive BBa_J23107 promoter and a gRNA module expressed from the strong constitutive BBa_J23119 promoter; the F plasmid origin of transfer (oriTF); the pBBR1 origin of replication (oriV), and a set of resistance cassettes (Ap, ampicillin; Kn, kanamycin; Cm, chloramphenicol; St, streptomycin; Gm, gentamycin), an optional cassette carrying the sfgfp or mcherry genes highly expressed from the broad-host range synthetic BioFab promoter. (B) Diagram of the TAP antibacterial strategy. A donor strain produces the F plasmid conjugation machinery to transfer the TAP into the recipient strain. Targeted recipient carries a sequence recognized by CRISPR(i) system that induces killing or gene expression inhibition. Non-targeted recipient lacking the spacer recognition sequence are insensitive to CRISPR(i) activity. (C) Histogram of TAPs transfer estimated by flow cytometry show that TAPkn-Cas9-nsp-GFP and TAPkn-Cas9-lacZ2-GFP are transferred with similar efficiency in recipient cells after 3 h of mating. Donors TAP-Cas9-nsp (LY1371) or TAP-Cas9-lacZ2 (LY1380), recipient HU-mCherry lac+ (LY248). Two-tailed unpaired t-test was performed. ns: non-significant P-value >0.05. (D) Histograms of the concentration of viable transconjugants estimated by plating assays show viability loss associated with the acquisition of TAP-Cas9-lacZ2. The corresponding percentage of viable transconjugants (ratio T/R+T) is shown above each bar. Two-tailed unpaired t-test was performed. **P-value <0.0021 (E) Fold-increase of the recipient population counts over the 3 h of mating. Donors TAP-Cas9-nsp (LY1369) or TAP-Cas9-lacZ2 (LY1370), recipient lac+ (LY827). Two-tailed unpaired t-test was performed. *P-value <0.05 (C–E) Mean and SD are calculated from three independent experiments.